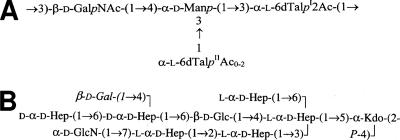Abstract
By the isolation of three different Aeromonas hydrophila strain AH-3 (serotype O34) mutants with an altered lipopolysaccharide (LPS) migration in gels, three genomic regions encompassing LPS core biosynthesis genes were identified and characterized. When possible, mutants were constructed using each gene from the three regions, containing seven, four, and two genes (regions 1 to 3, respectively). The mutant LPS core structures were elucidated by using mass spectrometry, methylation analysis, and comparison with the full core structure of an O-antigen-lacking AH-3 mutant previously established by us. Combining the gene sequence and complementation test data with the structural data and phenotypic characterization of the mutant LPSs enabled a presumptive assignment of all LPS core biosynthesis gene functions in A. hydrophila AH-3. The three regions and the genes contained are in complete agreement with the recently sequenced genome of A. hydrophila ATCC 7966. The functions of the A. hydrophila genes waaC in region 3 and waaF in region 2 were completely established, allowing the genome annotations of the two heptosyl transferase products not previously assigned. Having the functions of all genes involved with the LPS core biosynthesis and most corresponding single-gene mutants now allows experimental work on the role of the LPS core in the virulence of A. hydrophila.
In gram-negative bacteria, the lipopolysaccharide (LPS) is one of the major structural and immunodominant molecules of the outer membrane. It consists of three domains: lipid A, core oligosaccharide, and O-specific polysaccharide or O antigen. The genetics of O-antigen biosynthesis has been intensively studied in the Enterobacteriaceae and some other gram-negative bacteria. In studies of several Enterobacteriaceae, like Escherichia coli, Salmonella enterica, and Klebsiella pneumoniae, genes involved in LPS core biosynthesis are usually found clustered in a region of the chromosome, the waa gene cluster (13, 29). This gene arrangement is not always present in other gram-negative bacteria (28); e.g., in Bordetella species, the genes involved in the biosynthesis of the O antigen and core are present in the same gene cluster (2). On the other hand, a careful analysis of several fully sequenced genomes suggested that the genes for the LPS core biosynthesis may not be clustered and may be distributed between several regions, e.g., as in Yersinia pestis (24).
The overwhelming majority of the LPSs studied (15) contain at least one residue of 3-deoxy-d-manno-oct-2-ulosonic (ketodeoxyoctonic) acid (Kdo), which links the core to the lipid A moiety (KdoI). The second characteristic sugar of the core is l-glycero-d-manno-heptose (ld-Hep), although there are a few LPSs that contain d-glycero-d-manno-heptose (dd-Hep) or lack any heptose (8). In those containing ld-Hep, the presence of a Hep-α-(1→5)-Kdo disaccharide is a characteristic feature. In several gram-negative bacteria, like Haemophilus or Vibrio species, KdoI is phosphorylated at position 4 (11, 35), whereas in S. enterica, E. coli, and some others, KdoI is glycosylated at O-4 with a second Kdo residue (KdoII). Either KdoI (in Acinetobacter species) or KdoII (e.g., in Burkholderia cepacia and Yersinia pestis [15]) can be replaced with d-glycero-d-talo-oct-2-ulosonic acid (Ko). The biosynthesis pathway of the latter and regulation of its expression in the LPS remain unknown.
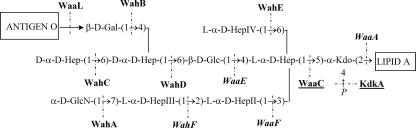
Mesophilic, motile Aeromonas species are opportunistic and primary pathogens of a variety of aquatic and terrestrial animals, including humans; the clinical manifestations range from gastroenteritis to soft-tissue infections, including septicemia and meningitis (4, 20). The varied clinical picture of Aeromonas infections and gastroenteric illness in particular suggests that complex pathogenic mechanisms are involved in this bacterium. Serotype O34 is most common among mesophilic Aeromonas spp. (19), accounting for 26.4% of all infections. Previous investigations have documented O34 strains as important causes of infections in humans (18, 19). Recently, we have established the structures of the O34 antigen (22) and the LPS core of Aeromonas hydrophila AH-901 (23), an O-antigen-lacking mutant derived from a typical wild-type strain, AH-3, belonging to serotype O34 (Fig. 1).
FIG. 1.
Chemical structures of O34-antigen LPS (A) and the LPS core (B) of A. hydrophila strain AH-3 (22, 23). O34-antigen LPS is linked to the Gal residue (shown in italics) of the LPS core (23).
Now we report the identification and characterization of three genomic regions in A. hydrophila AH-3 (serotype O34) involved in the LPS core biosynthesis. Combining these data together with the structure elucidation of the LPS core in mutants in each gene from the three gene clusters enabled a presumptive assignment of all LPS core biosynthesis gene functions.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Aeromonas strains were routinely grown in tryptic soy broth (TSB) or on tryptic soy agar at 30°C unless stated otherwise. E. coli strains were grown on Luria-Bertani (LB) Miller broth and LB Miller agar at 37°C. Kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), tetracycline (20 μg ml−1), rifampin (100 μg ml−1), or chloramphenicol (25 μg ml−1) was added to the different media.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F−endA hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA96 φ80lacZM15 | 12 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F−proAB lacIqZΔM15 Tn10) | Stratagene |
| S17-1 | hsdR pro recA, RP4-2 in chromosome Km::Tn7 (Tc::Mu) | 7 |
| MC1061 | thi thr-1 leu6 proA2 his-4 argE2 lacY1 galK2 ara-14 xyl-5 supE44 λpir | 31 |
| A. hydrophila | ||
| AH-3 | O34; wild type | 25 |
| AH-405 | AH-3; spontaneous Rifr | 25 |
| AH-3005 | AH-3 ORF6.1:mini-Tn5Km-1; Rifr Kmr | This study |
| AH-3006 | AH-3 ORF3.2:mini-Tn5Km-1; Rifr Kmr | This study |
| AH-3007 | AH-3 ORF1.3:mini-Tn5Km-1; Rifr Kmr | This study |
| AH-3Δ2.1 | AH-3 ORF2.1 mutant in frame with pDM4 | This study |
| AH-3Δ3.1 | AH-3 ORF3.1 mutant in frame with pDM4 | This study |
| AH-3Δ4.1 | AH-3 ORF4.1 mutant in frame with pDM4 | This study |
| AH-3Δ5.1 | AH-3 ORF5.1 mutant in frame with pDM4 | This study |
| AH-3Δ2.2 | AH-3 ORF2.2 mutant in frame with pDM4 | This study |
| AH-3Δ7.1 | AH-3 ORF7.1 insertion mutant with pFS100; Kmr | This study |
| AH-3Δ4.2 | AH-3 ORF4.2 insertion mutant with pFS100; Kmr | This study |
| Plasmids | ||
| pRK2073 | Helper plasmid; Spcr | 7 |
| pLA2917 | Cosmid vector; Kmr Tcr | 27 |
| COS-CORE2 | pLA2917 cosmid with AH-3 core region 2; Tcr | This study |
| COS-CORE3 | pLA2917 cosmid with AH-3 core region 3; Tcr | This study |
| pGEM-T | PCR cloning vector; Apr | Promega |
| pGEMT-ORF2.3 | pGEM-T with AH-3 ORF2.3; Apr | This study |
| pGEMT-ORF2.3-1.2 | pGEM-T with AH-3 ORF2.3 and ORF1.2; Apr | This study |
| pDM4 | pir dependent with sacAB genes, oriR6K; Cmr | 26 |
| pDM4Δ2.1 | pDM4 with AH-3ΔORF2.1; Cmr | This study |
| pDM4Δ3.1 | pDM4 with AH-3ΔORF3.1; Cmr | This study |
| pDM4Δ4.1 | pDM4 with AH-3ΔORF4.1; Cmr | This study |
| pDM4Δ5.1 | pDM4 with AH-3ΔORF5.1; Cmr | This study |
| pDM4Δ2.2 | pDM4 with AH-3ΔORF2.2; Cmr | This study |
| pFS100 | pGP704 suicide plasmid, pir dependent; Kmr | 31 |
| pFS-7.1 | pFS100 with an internal fragment of ORF7.1; Kmr | This study |
| pFS-4.2 | pFS100 with an internal fragment of ORF4.2; Kmr | This study |
| pBAD33 | Arabinose-inducible expression vector | ATCC |
| pABD-ORF | pBAD33 with the corresponding ORF; Cmr | This study |
Rif, rifampin; Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline; Ap, ampicillin; Spc, spectinomycin.
Mini-Tn5Km-1 mutagenesis.
The conjugal transfer of transposition element mini-Tn5Km-1 from E. coli S17-1 λpirKm-1 (7) to A. hydrophila AH-405 (AH-3 rifampin resistant) was carried out in a conjugal drop incubated for 6 h at 30°C in 1:5:1 ratios of S17-1 λpirKm-1 to AH-405 to HB101/pRK2073 (helper plasmid), respectively. The serial dilutions of the mating mix were plated on tryptic soy agar supplemented with rifampin and kanamycin in order to select mutants.
General DNA methods.
General DNA manipulations were done essentially as previously described (32). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers.
DNA sequencing and computer analysis of sequence data.
Double-stranded DNA sequencing was performed by using the dideoxy-chain termination method (33) with the ABI Prism dye terminator cycle sequencing kit (Perkin Elmer). Oligonucleotides used for genomic DNA amplifications and DNA sequencing were purchased from Pharmacia LKB Biotechnology. The DNA sequence was translated in all six frames, and all open reading frames (ORFs) were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST (3, 5) network service at the National Center for Biotechnology Information and the European Biotechnology Information, respectively. ClustalW was used for multiple-sequence alignments.
Southern and dot blot hybridizations.
Southern blotting was performed by capillary transfer (32). For dot blot hybridizations, the DNA was denatured by boiling for 5 min, chilled on ice for another 5 min, and spotted onto Hybond-N1 (Amersham) nylon membrane. Probe labeling, hybridization, and detection were carried out by using the enhanced chemiluminescence labeling and detection system (Amersham) according to the manufacturer's instructions.
Mutant construction.
The chromosomal in-frame AH-3Δ2.1, AH-3Δ3.1, AH-3Δ4.1, AH-3Δ5.1, and AH-3Δ2.2 deletion mutants were constructed by allelic exchange as described by Milton et al. (26). The primers used to obtain the mutants are listed in Table SP1 in the supplemental material. Two asymmetric PCRs were carried out to obtain two DNA fragments (A-B and C-D) that were annealed at their overlapping regions and PCR amplified as a single DNA fragment using primers A and D. The amplified in-frame deletion gene products were purified, SalI or BglII digested, ligated into SalI- or BglII-digested and phosphatase-treated pDM4 vector, electroporated into E. coli MC1061 (λpir), and plated on chloramphenicol LB agar plates at 30°C to obtain the pDM4 plasmids with the corresponding deleted genes. Each mutated gene was transferred to the chromosome by homologous recombination using the λpir-dependent suicide plasmid pDM4, which contained the counterselectable marker sacB. Triparental mating with the mobilizing strain HB101/pRK2073 was used to transfer the plasmids containing the engineered in-frame deletions into the A. hydrophila AH-405 rifampin-resistant strain. Transconjugants were selected on plates containing chloramphenicol and rifampin at 30°C. PCR analysis confirmed that the vector had integrated correctly into the chromosomal DNA. To complete the allelic exchange, the integrated suicide plasmid was forced to recombine out of the chromosome by growth on agar plates containing 15% sucrose. Mutants were selected based on their survival on plates containing 15% sucrose and the loss of the chloramphenicol-resistant marker of vector pDM4. The mutations were confirmed by sequencing the whole constructs in amplified PCR products.
Mutants AH-3Δ7.1 and AH-3Δ4.2 were obtained as defined insertion mutants by the integration of suicide plasmid pFS100 (31), carrying an internal fragment from the gene. Primers and restriction sites used are indicated in Table SP1 in the supplemental material. Mutants AH-3Δ6.1 (AH-3005), AH-3Δ3.2 (AH-3006), and AH-3Δ1.3 (AH-3007) were obtained by mini-Tn5 insertion.
Plasmid constructions and mutant complementation studies.
For complementation studies, the A. hydrophila AH-3 genes (ORF2.1, ORF3.1, ORF4.1, ORF5.1, ORF6.1, ORF7.1, ORF2.2, ORF3.2, ORF4.2, and ORF1.3) were PCR amplified by using specific primer pairs and ligated to plasmid pBAD33 (see the list of primers in Table SP2 in the supplemental material). The plasmid constructs were transformed into E. coli LMG194 by electroporation, plated on chloramphenicol LB agar plates, and incubated at 30°C. Plasmids with the amplified genes were independently transferred into the corresponding mutants by triparental mating by using the mobilizing strain HB101/pRK2073. Transconjugants were selected on plates containing chloramphenicol and rifampin and confirmed by PCR. Each gene was expressed from the arabinose-inducible and glucose-repressible pBAD33 promoter. Repression from the araC promoter was achieved by growth in medium containing 0.2% (wt/vol) d-glucose, and induction was obtained by adding l-arabinose to a final concentration of 0.2% (wt/vol). The cultures were grown for 18 h at 30°C in TSB medium supplemented with chloramphenicol and 0.2% glucose, diluted 1:100 in fresh medium (without glucose), and grown until they reached an A600 of about 0.2. Then, l-arabinose was added, and the cultures were grown for another 2 h. Repressed controls were maintained in glucose-containing medium.
To obtain the pGEMT-ORF2.3-1.2 plasmid, the A. hydrophila AH-3 kdkA (ORF2.3) gene and its putative promoter site were amplified by PCR using chromosomal DNA with primers 2.3-F (5′-GTGACAACAATCCCCGATG-3′) and 2.3-R (5′-ATCAGCGCCAGATCAAACT-3′). The amplified DNA fragment (1,099 bp) was ligated into the vector pGEM-T (Promega) to obtain the pGEMT-ORF2.3 plasmid and transformed into E. coli XL1-Blue. Transformants were selected on LB plates containing ampicillin. The A. hydrophila AH-3 waaA (ORF1.2) gene was PCR amplified from chromosomal DNA with primers 1.2-F (5′-ACGCGTCGACCCGATCGTGCTGCAAGTG-3′) and 1.2-R (5′-ACGCGTCGACCACGACCTTCAGCGACTC-3′). The PCR product (1,519 bp) was digested with SalI (sites underlined above) and ligated to the SalI-digested and phosphatase-treated pGEMT-ORF2.3 to generate the pGEMT-ORF2.3-1.2 plasmid, which was transformed into E. coli XL1-Blue.
LPS isolation and SDS-PAGE.
For LPS analysis, cultures were grown in TSB at 20 or 37°C. LPS was isolated by the method of Galanos et al. (10), resulting in a 2.3% yield. For screening purposes, LPS was obtained after proteinase K digestion of whole cells (9). LPS samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or SDS-Tricine-PAGE and visualized by silver staining as previously described (9, 14).
Large-scale isolation and mild-acid degradation of LPS.
Dry bacterial cells of each mutant in 25 mM Tris-HCl buffer containing 2 mM CaCl2, pH 7.63 (10 ml g−1), were treated at 37°C with RNase, with DNase (24 h, 1 mg g−1 each), and then with proteinase K (36 h, 1 mg g−1). The suspension was dialyzed and lyophilized, and the LPS was extracted by the phenol-water procedure (36).
A portion of the LPS (∼50 mg) from each strain was heated with aqueous 2% acetic acid (6 ml) at 100°C for 45 min. The precipitate was removed by centrifugation (13,000 × g, 20 min) and the supernatant was fractionated on a column (56 by 2.6 cm) of Sephadex G-50 in 0.05 M pyridinium acetate buffer, pH 4.5, with monitoring by using a differential refractometer (Knauer, Germany). An oligosaccharide fraction was obtained in a yield of 9 to 20%, depending on the strain.
Methylation analysis.
A sample of each oligosaccharide (0.5 mg) was dissolved in 1 ml dimethyl sulfoxide. An excess of powdered NaOH was added, and the reaction glass was flushed with dry N2 and sealed. After the mixture was stirred at 20°C for 1 h, 0.5 ml of cold CH3I was added and the mixture was stirred at 20°C for 1 h. Then, water was added, the methylated product was extracted with CHCl3, and the extract was washed with water and evaporated with a stream of dry nitrogen. The methylated oligosaccharide was hydrolyzed with 2 M CF3CO2H (120°C, 2 h), and acid was removed with a stream of nitrogen. The methylated monosaccharides were conventionally reduced with NaBH4, acetylated with acetic oxide in pyridine, and analyzed by gas-liquid chromatography (GLC) on a Hewlett-Packard 5880 chromatograph and by GLC-mass spectrometry (MS) on a Hewlett-Packard HP 5989A instrument using an HP-5ms capillary column and a temperature gradient of 150°C (3 min) to 320°C with an increase of 5°C min−1.
MS.
Negative-ion electrospray ionization with Fourier transform ion cyclotron resonance MS was performed on an APEX II instrument (Bruker Daltonics) equipped with an actively shielded 7-T magnet and an Apollo ion source. Mass spectra were acquired by using standard experimental sequences as provided by the manufacturer. The mass scale was calibrated externally with rough LPS of known structure. Samples (∼10 ng μl−1) were dissolved in 50:50:0.001 (vol/vol/vol) mixtures of 2-propanol, water, and triethylamine with a pH of ∼8.5 and sprayed at a flow rate of 2 μl min−1. The capillary entrance voltage was set to 3.8 kV, and the drying gas temperature was set to 150°C. The capillary exit voltage was set to −100 V and in some cases was increased to −200 V to get a better signal intensity. The spectra showing several charge states for each component were charge deconvoluted using Bruker XMASS 6.0.0 software, and the mass numbers given refer to monoisotopic molecular masses.
Nucleotide sequence accession numbers.
The complete nucleotide sequences of the three A. hydrophila AH-3 chromosomal regions containing LPS core biosynthesis genes described here have been assigned the following GenBank accession numbers: EU296246, EU296247, and EU296248.
RESULTS
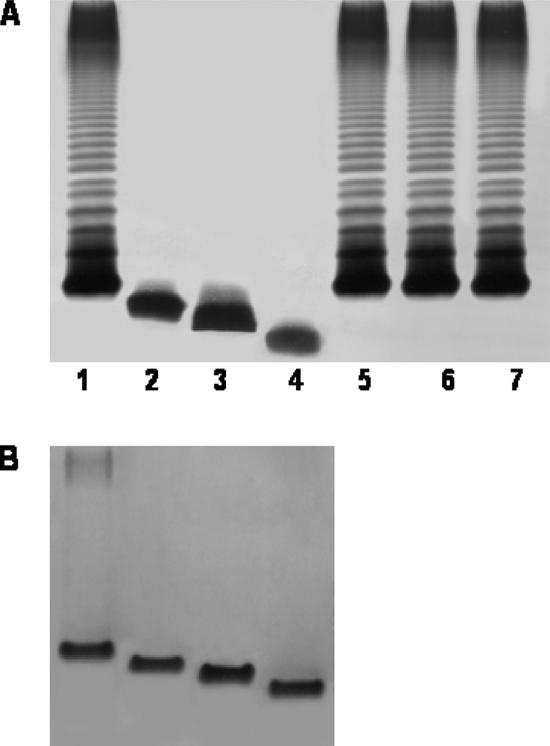
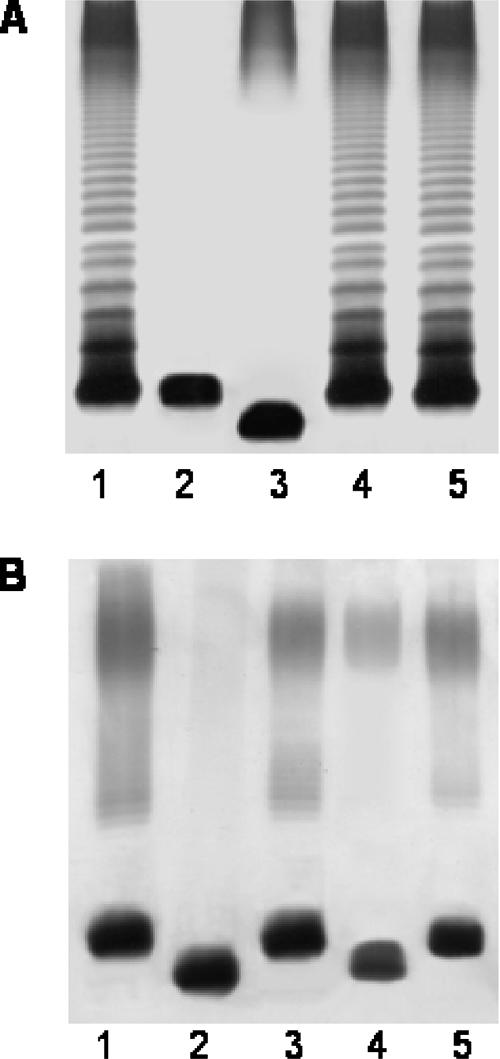
After the mutagenesis of a rifampin-resistant isolate (AH-405) of the A. hydrophila wild-type strain AH-3 (serotype O34) (25), mutants (Kmr) were screened for their inability to react with specific serum against O34-antigen LPS (7). Mutants AH-3005, AH-3006, and AH-3007 were among 3,000 mutants that were initially screened (1.7% serum-resistant mutants) and showed a lack of O34 antigen and a faster mobility of R-type LPS (lipid A and core) in gel (Fig. 2). The AH-3007 LPS moved faster than that of AH-3006, which moved faster than the AH-3005 LPS, thus indicating that genes implicated in the biosynthesis of the AH-3 LPS core could be affected by the transposon. Southern blot analysis using a specific probe for the transposon demonstrated that each mutant had a single copy of a minitransposon in the genome (data not shown). After the cloning of the minitransposon containing a fragment from flanking DNA from the genome of each mutant (EcoRV digestion) in pBCSK (Stratagene), sequence analysis was performed using the oligonucleotides from the mini-Tn5Km-1 flanking regions, 5′-AGATCTGATCAAGAGACAG-3′ and 5′-ACTTGTGTATAAGAGTCAG-3′, and the universal oligonucleotides from the plasmid vector, T3 and T7. Three different ORFs (one for each mutant) encoding proteins, all similar to heptosyl transferases from different bacteria, were identified. These sequence data were used to synthesize three internal DNA probes from the A. hydrophila AH-3 genome (named 1, 2, and 3 for the heptosyl transferases of AH-3005, AH-3006, and AH-3007, respectively).
FIG. 2.
(A) LPS samples were extracted and analyzed by SDS-PAGE (12%) as described by Darveau and Hancock (9). Results for SDS-PAGE-extracted LPSs from A. hydrophila AH-3 (wild type; lane 1), mutants AH-3005, AH-3006, and AH-3007 (lanes 2, 3, and 4, respectively), and mutants complemented with plasmids pBAD-ORF6.1, pBAD-ORF3.2, and pBAD-ORF1.3 (lanes 5, 6, and 7, respectively) are shown. (B) SDS-Tricine-PAGE gels from analysis of the LPS cores of mesophilic Aeromonas strains (lanes 1 to 4, same as for panel A). The strains with pBAD33 plasmids were grown under induced conditions.
Cloning and sequencing of the three A. hydrophila AH-3 genomic regions comprising the waa genes in E. coli K-12 strains.
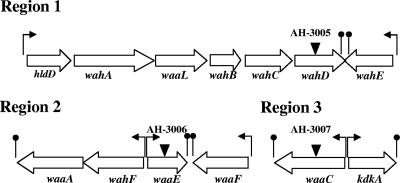
A. hydrophila AH-3 is a typical strain of serotype O34 (25). A cosmid-based genomic library of A. hydrophila AH-3 was constructed and introduced into E. coli DH5α (27). Tetracycline-resistant clones were screened by colony blotting using the DNA probes previously obtained from the regions close to the mini-Tn5 location in the different mutant strains. An independent recombinant positive clone was found for probes 2 and 3, named COS-CORE2 and COS-CORE3, respectively. No recombinant positive clone was obtained for probe 1 after extensive analysis of either more than 5,000 clones or a clone able to hybridize with more than one probe. With the genome DNA region of probe 1, the nucleotide sequence was spanned, either downstream or upstream of the region, by genome-walking PCR (37). The analysis of sequenced region 1 showed seven genes involved in the LPS core biosynthesis (Fig. 3), and the corresponding analysis of the proteins encoded by these ORFs with their homologies is presented in Table 2.
FIG. 3.
Genetic organization of three A. hydrophila AH-3 chromosomal regions containing genes for LPS core biosynthesis. Regions 2 and 3 are from COS-CORE2 and COS-CORE3 plasmids, respectively. Transcription direction and stops (lollipops) are indicated. The locations of mini-Tn5 insertions (for AH-3005, AH-3006, and AH-3007) are shown.
TABLE 2.
Characteristics of the three A. hydrophila AH-3 regions containing genes for LPS core biosynthesis
| Region | ORF
|
Nucleotide position | Protein size (no. of amino acids) | Homologous protein (species) | Accession no. | % Identity/% similarity | |
|---|---|---|---|---|---|---|---|
| No. | Protein encoded | ||||||
| 1 | 1 | HldD | 591-1550 | 319 | ADP-l-glycero-d-manno-heptose 6-epimerase (Klebsiella pneumoniae) | Q9XCA1 | 71/83 |
| 2 | WahA | 1601-3358 | 586 | Glycosyltransferase (WavL) (Vibrio cholerae) | AAL77362 | 64/77 | |
| 3 | WaaL | 3368-4492 | 374 | Lipid-A-core, O-antigen ligase from different bacteria | ZP_00202015 | 23/45 | |
| 4 | WahB | 4577-5242 | 221 | Glycosyltransferase (Actinobacillus pleuropneumoniae) | AAD30156 | 50/68 | |
| 5 | WahC | 5328-6356 | 342 | ADP-heptose-LPS heptosyltransferase, either I, II, or III from different bacteria | YP_109261 | 28/42 | |
| 6 | WahD | 6411-7520 | 370 | ADP-heptose-LPS heptosyltransferase (Actinobacillus pleuropneumoniae) | AAD30157 | 26/48 | |
| 7 | WahE | 8526-7462 | 355 | ADP-heptose-LPS heptosyltransferase (Fusobacterium nucleatum) | ZP_00145125 | 27/54 | |
| 2 | 1 | WaaA | 1585-320 | 421 | 3-deoxy-d-manno-octulosonic acid transferase (Vibrio cholerae and other species) | AAF93409 | 53/71 |
| 2 | WahF | 2741-1587 | 384 | ADP-heptose-LPS heptosyltransferase II (Vibrio parahaemolyticus) | NP_796591 | 57/73 | |
| 3 | WaaE | 2944-3732 | 262 | α-l-glycero-d-manno-heptose β-1,4-glucosyltransferase (Vibrio fischeri) | YP_203517 | 56/70 | |
| 4 | WaaF | 4837-3794 | 347 | Probable ADP-heptose-LPS heptosyltransferase II (Vibrio cholerae and other species) | AAF93399 | 59/73 | |
| 3 | 1 | WaaC | 1476-421 | 351 | ADP-heptose-LPS heptosyltransferase (Vibrio cholerae and other species) | ZP_00757651 | 58/73 |
| 2 | KdkA | 1661-2374 | 273 | 3-deoxy-d-manno-octulosonic acid kinase (Vibrio alginolyticus and other species) | ZP_01261295 | 44/65 | |
The nucleotide sequences of the DNA inserts of plasmids COS-CORE2 and COS-CORE3 were determined in order to identify the A. hydrophila AH-3 genes conferring the LPS core production. The complete nucleotide sequence was determined in both directions by using oligonucleotides complementary to cosmid pLA2917 sequences flanking the COS-CORE2 and COS-CORE3 DNA inserts. Other sequence-derived oligonucleotides were purchased (Amersham-Pharmacia Biotech) and used to complete the nucleotide sequence. The analysis of the sequenced regions showed genes involved in LPS core biosynthesis in both regions (named 2 and 3 for COS-CORE2 and COS-CORE3, respectively), as indicated in Fig. 3. The corresponding analysis of the proteins encoded by these ORFs in both regions with their homologies is presented in Table 2. All the homologies in Table 2 included only proteins with confirmed functions.
Mutant isolation, phenotypic characterization, and complementation.
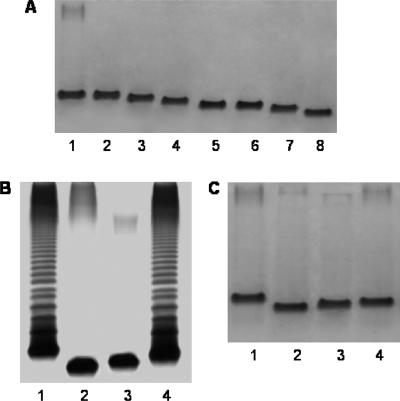
Mutants were named according to the ORF number and the region number; for instance, AH-3Δ2.1 indicates a mutant with a mutation in ORF2 of region 1. As judged by using SDS-PAGE, mutants AH-3Δ3.1, AH-3Δ4.1, AH-3Δ6.1, AH-3Δ2.2, AH-3Δ3.2, AH-3Δ4.2, and AH-3Δ1.3 completely lacked O34-antigen LPS, and R-type LPS from all mutants but AH-3Δ3.1 (a putative waaL mutant) migrated faster to different degrees than the R-type LPS of the wild-type strain (Fig. 4A). Mutants AH-3Δ2.1, AH-3Δ5.1, and AH-3Δ7.1 showed different amounts of O34-antigen LPS in gels, but some changes in the migration of the R-type LPS compared to that of the wild-type LPS were observed (Fig. 4B and C). No AH-3Δ1.2 or AH-3Δ2.3 mutants were obtained for two genes similar to those obtained for Kdo transferase (waaA) and Kdo kinase (kdkA).
FIG. 4.
(A) SDS-Tricine-PAGE gels for the wild type, rifampin-resistant strain AH-405 (lane 1), and the LPS core mutants lacking O34-antigen LPS, namely, AH-3Δ3.1, AH-3Δ4.1, AH-3Δ6.1, AH-3Δ2.2, AH-3Δ3.2, AH-3Δ4.2, and AH-3Δ1.3 (lanes 2 to 8, respectively). SDS-PAGE (B) and SDS-Tricine-PAGE (C) gels of the wild type, rifampin-resistant strain AH-405 (lane 1), and the LPS core mutants showing different amounts of O34-antigen LPS, namely, AH-3Δ2.1, AH-3Δ5.1, and AH-3Δ7.1 (lanes 2 to 4, respectively).
Complementation with the corresponding wild-type gene expressed in the vector plasmid pBAD33 restored the complete LPS in all mutants, as revealed by SDS-PAGE. No such complementation was achieved with the plasmid vector alone. As an example, Fig. 5A shows the complementation of AH-3Δ3.1 and AH-3Δ2.1 mutants with plasmids pBAD-ORF3.1 and pBAD-ORF2.1, respectively. In addition, plasmid pBAD-ORF3.2 was fully able to complement the Klebsiella pneumoniae 52145ΔwaaE mutant (16), and plasmid pBAD-ORF4.2 was the only one carrying different putative heptosyltransferase genes able to complement the K. pneumoniae 52145ΔwaaF mutant (17) (Fig. 5B). None of the pBAD plasmid vectors carrying different putative heptosyltransferases was able to complement K. pneumoniae 52145ΔwaaC or E. coli waaC mutants (17). A similar result was obtained for the K. pneumoniae mutant 52145ΔwaaQ.
FIG. 5.
SDS-PAGE gels of LPSs from strains of A. hydrophila (A) and Klebsiella pneumoniae (B). (A) Lanes: 1, AH-405 (wild type); 2, AH-3Δ3.1 mutant; 3, AH-3Δ2.1 mutant; 4, AH-3Δ3.1 complemented with the pBAD-ORF3.1 plasmid; and 5, AH-3Δ2.1 complemented with the pBAD-ORF2.1 plasmid. (B) Lanes: 1, 52145 (O1:K2, wild type); 2, 52145ΔwaaF mutant (13); 3, 52145ΔwaaF complemented with the pBAD-ORF4.2 plasmid; 4, 52145ΔwaaE mutant (12); and 5, 52145ΔwaaE complemented with the pBAD-ORF3.2 plasmid. The strains with pBAD33 plasmids were grown under induced conditions.
E. coli strain CJB26 harbors a kanamycin determinant inserted in the waaA gene and its wild-type waaA gene in a temperature-sensitive plasmid (pJSC2), leading to a growth-temperature-sensitive phenotype (6). Plasmid pGEMT-1.2 (carrying ORF1.2 alone) was unable to complement E. coli CJB26 (it did not change the growth-temperature-sensitive phenotype), whereas plasmid pGEMT-ORF2.3-1.2 (carrying ORF1.2 and ORF2.3 together) was fully able to abolish the growth-temperature-sensitive phenotype. This experiment strongly suggests that ORF1.2 and ORF2.3 encode the Kdo transferase WaaA and the Kdo kinase KdkA of the A. hydrophila LPS inner-core biosynthesis pathway.
Structure elucidation of the mutant LPSs.
The LPSs were isolated by the phenol-water extraction of the enzymatically digested cells of each mutant. The LPS samples were degraded with a mild acid and the released core oligosaccharides were isolated by using gel permeation chromatography on Sephadex G-50. They were studied by high-resolution electrospray ionization MS and, when necessary, methylation analysis. The latter method enabled not only the determination of the linkage positions in most sugar residues but also differentiation between dd-Hep and ld-Hep residues, whose derivatives have different retention times in GLC.
AH-3Δ3.1 and AH-3Δ4.1.
The oligosaccharide sample from the first mutant was found to be essentially identical to that from the R-type LPS of A. hydrophila AH-901 (20). The major molecular ion peak at m/z 1,857.63 in its mass spectrum (see Figure SA in the supplemental material) corresponded to the full core (calculated molecular mass, 1,857.61 atomic mass units), whose structure is shown in Fig. 6. The mutant AH-3Δ4.1 showed a structure identical (Fig. 6) to that of the AH-3Δ3.1 mutant but lacked the Gal residue (major molecular ion peak at m/z 1,695.57).
FIG. 6.
Proposed structures of the core oligosaccharides released by mild-acid hydrolysis from the LPSs of the A. hydrophila mutants. When several oligosaccharides were obtained, components present in a nonstoichiometric quantity are shown in italics and the major glycoform is shown in bold. The assigned names of the mutated genes in this work are shown in parentheses.
AH-3Δ7.1 and AH-3Δ5.1.
The mass spectrum of the core from AH-3Δ7.1 (see Figure SA in the supplemental material) was similar to that of AH-3Δ3.1, but the masses were lower by 192 atomic mass units, which corresponded to the lack of a heptose residue. Methylation analysis showed that the lost residue was the terminal ld-HepIV, whereas the terminal dd-HepII was retained. In contrast, in AH-3Δ5.1, the terminal ld-HepIV was conserved and the terminal dd-HepII was lost. Based on the mass spectrum (see Figure SA in the supplemental material), the major core glycoform in AH-3Δ5.1 was also devoid of galactose (molecular ion peak at m/z 1,503.53). Therefore, the core oligosaccharides from these two mutants have the structures shown in Fig. 6.
AH-3Δ2.1.
The core from AH-3Δ2.1 was found to lack GlcN. The major glycoforms were also devoid of Gal and either one or three Hep residues (the latter is shown as a major glycoform in Fig. 6), as determined from the appearance in the mass spectrum of intense molecular ion peaks at m/z 1,342.43 (see Fig. SA in the supplemental material) and 958.30 (not shown), respectively.
AH-3Δ6.1.
For the AH-3Δ6.1 mutant, the mass spectrum showed the lack of the outer-core trisaccharide fragment consisting of Gal, dd-HepII, and dd-HepI (see Fig. SA in the supplemental material). Methylation analysis confirmed the conservation of the terminal ld-HepIV, the absence of Gal and any dd-Hep, and the appearance of the terminal Glc.
AH-3Δ2.2 and AH-3Δ3.2.
The cores from the AH-3Δ2.2 and AH-3Δ3.2 mutants showed similar mass spectra (see Fig. SA in the supplemental material). Both contained only one molecular ion peak at m/z 796.24 or 796.25 for an oligosaccharide consisting of one Kdo and three heptose residues. Methylation analysis of the former showed the presence of only terminal ld-Hep, whereas both terminal and 2-substituted ld-Hep residues were revealed in the latter. Based on these data, the branched and linear structures were assigned to the oligosaccharides from AH-3Δ2.2 and AH-3Δ3.2, respectively (Fig. 6).
AH-3Δ4.2.
For AH-3Δ4.2, a further-truncated core containing one Kdo and two heptose residues was identified by the presence in the mass spectrum of an intense molecular ion peak at m/z 604.19 (see Fig. SA in the supplemental material).
AH-3Δ1.3.
As the LPS from the AH-3Δ1.3 mutant gave no oligosaccharide upon mild-acid degradation, it was studied as a whole. The mass spectrum (see Fig. SB in the supplemental material) showed that the core region is restricted to a Kdo phosphate.
DISCUSSION
The isolation by transposon mutagenesis of three different mutants from A. hydrophila AH-3 devoid of O34-antigen LPS and altered R-type LPS migration in gels allowed the characterization of three different genomic regions with LPS core biosynthesis genes. Region 1 contained seven genes flanked by genes encoding a putative regulator, TetR, and a hypothetical protein, region 2 contained four genes flanked by genes encoding an integral membrane protein of unknown function and CoaD, and region 3 contained two genes flanked by genes encoding a guanylate kinase and a protein with a GGDEF domain. The three regions and the genes contained are in complete agreement with those recently sequenced for A. hydrophila ATCC 7966 (34), in which region 1 (AHA4232 to AHA4226), region 2 (AHA0170 to AHA0167), and region 3 (AHA0042 to AHA0043) are flanked by the same putative genes. While the chemical structure of the A. hydrophila AH-3 LPS is already known (22, 23), this is not the case for A. hydrophila ATCC 7966. The presumptive assignment of all genes involved in the LPS core biosynthesis of A. hydrophila AH-3, as well as the distribution of inner- and outer-core biosynthesis genes between regions 1, 2, and 3, is shown in Fig. 7.
FIG. 7.
Complete presumable assignment of the genes involved in the LPS core biosynthesis of A. hydrophila AH-3. Proteins encoded by genes from different regions are shown in roman (region 1), italic (region 2), or underlined (region 3) type.
Region 3.
Two genes of region 3, ORF1 and ORF2, encode ld-HepI transferase WaaC and Kdo kinase KdkA, respectively. The function of the waaC gene was confirmed by the mass spectrum of the whole LPS of the mutant AH-3Δ1.3. The A. hydrophila AH-3 LPS consists of a phosphorylated Kdo residue (most likely, Kdo 4-phosphate) as in Vibrio spp. (23, 35), which is in agreement with the similarity between A. hydrophila WaaC and a heptosyl transferase of Vibrio (not determined to be WaaC). This also accounts for the failure of A. hydrophila WaaC complementation in Enterobacteriaceae, which possess a nonphosphorylated Kdo in the core. Furthermore, the failure of isolation of a kdkA mutant could be due to the lethality of a mutation in this gene encoding Kdo kinase.
Region 2.
Region 2 includes four genes, ORF1 to ORF4, which encode Kdo transferase WaaA, a new heptosyl transferase that we named WahF, glucosyl transferase WaaE, and ld-HepII transferase WaaF, respectively. Mutation in the A. hydrophila gene waaA seems to be lethal, as it has been reported for the majority of gram-negative bacteria (28). In E. coli, Kdo is not phosphorylated. A. hydrophila WaaA is able to complement E. coli strain CJB26 (a temperature-sensitive waaA mutant) only when coexpressed with the A. hydrophila Kdo kinase KdkA, thus confirming the functions of both proteins. WaaE is l-glycero-d-manno-heptose β-1,4-d-glucosyltransferase not only by homology but also by its ability to complement the K. pneumoniae 52145ΔwaaE mutant. The core oligosaccharide in the waaE mutant AH-3Δ3.2 represents a remaining linear fragment of the core consisting of Kdo and three ld-Hep residues (Fig. 6). This result suggests that the transfer of the Glc residue precedes the incorporations of the outer core, GlcN, and ld-HepIV residues. ORF4 encodes ld-HepII transferase WaaF, as this is the only heptosyltransferase able to fully complement the K. pneumoniae 52145ΔwaaF mutant. Its function was confirmed by the mass spectrum of the core oligosaccharide from the waaF mutant AH-3Δ4.2, which showed a further-truncated core containing only Kdo and two ld-Hep residues (Fig. 6). Hence, the incorporation of ld-HepII precedes that of Glc. WahF was assigned as l-glycero-d-manno-heptose α-1,2-l-glycero-d-manno-heptosyltransferase (ld-HepIII transferase) since the corresponding mutant, AH-3Δ2.2, has a truncated core lacking ld-HepIII but containing the three other ld-Hep residues, as shown in Fig. 6. Accordingly, WahF was unable to complement the K. pneumoniae 52145ΔwaaF mutant, and an A. hydrophila AH-3 waaE wahF double mutant has a trisaccharide LPS core consisting of Kdo, ld-HepI, and ld-HepII (data not shown).
Region 1.
From seven genes present in region 1, ORF1 was identified as ADP-d-glycero-β-d-manno-heptose-6-epimerase due to the fact that an extremely high homology was observed. This enzyme converts the dd-Hep derivative into the corresponding ld-Hep derivative (21). ORF2 was named WahA and assigned as bifunctional l-glycero-d-manno-heptose α-1,7-N-acetylglucosaminyltransferase/N-deacetylase. The WahA protein showed two domains: a glycosyltransferase group 1 domain (amino acid residues 156 to 303) and a polysaccharide deacetylase domain (amino acid residues 404 to 543). The core from the wahA mutant AH-3Δ2.1 lacked GlcN but an oligosaccharide with all other core components present was minor, whereas the major oligosaccharides were also devoid of a part of the outer core or the whole outer core and one of the ld-Hep residues. The AH-3Δ2.1 mutant LPS still showed some O34 antigen in gel, but it was a smaller amount than the O34 antigen of the wild-type strain.
ORF3 was assigned as WaaL since the waaL mutant AH-3Δ3.1 produced an LPS with the full core but was completely devoid of the O34 antigen. As in the wild-type strain, the terminal Gal was present in a nonstoichiometric amount. WaaL showed 11 transmembrane helices between amino acid residues 15 and 31, 40 and 56, 69 and 85, 90 and 106, 113 and 132, 149 and 165, 178 and 194, 199 and 215, 222 and 238, 331 and 347, and 354 and 370, as is characteristic for lipid-A-core, O-antigen ligases (1). The ORF4 product was named WahB and designated d-glycero-d-manno-heptose β-1,4-galactosyltransferase based on the fact that the LPS of the wahB mutant AH-3Δ4.1 lacks only galactose from the core and the O34 antigen. The ORF5 product, named WahC, was designated d-glycero-d-manno-heptose α-1,6-d-glycero-d-manno-heptosyltransferase, as the core from the wahC mutant AH-3Δ5.1 was devoid of dd-HepII. The presence of dd-HepII seems to be important for the incorporation of Gal into the core since only a minor amount of Gal and, consequently, a reduced amount of the O34 antigen were found in the mutant LPS.
The ORF6 product, named WahD, was assigned as d-glucose α-1,6-d-glycero-d-manno-heptosyltransferase (dd-HepI transferase) based on the lack of O34 antigen and the outer core from the LPS of the corresponding mutant AH-3Δ6.1. The ORF7 product was named WahE and assigned as l-glycero-d-manno-heptose α-1,6-l-glycero-d-manno-heptosyltransferase (ld-HepIV transferase) since the core from the wahE mutant differed from the full core only in its lack of ld-HepIV. This assignment is consistent with the presence of the O34 antigen in the mutant AH-3Δ7.1.
The data obtained show that the full inner core consisting of Kdo, three ld-Heps (I, II, and III), and Glc, i.e., the LPS domain synthesized by inner-core gene regions 2 and 3, is absolutely necessary for the addition of the outer-core monosaccharides. The absence of GlcN significantly affected the incorporation of the outer-core sugars, too. While the contributions of lipid A and O antigen to pathogenesis have been well studied, so far, little is known about the role of the LPS core domain. The LPS core domain seems to contribute to pathogenesis or to adaptation to several body sites, as suggested by the prevalence of some core types within clinical isolates. The interchanging of two core types of K. pneumoniae has a measurable effect on its virulence in an animal model of experimental infection (30). Mutation in either of the genes for biosynthesis of the inner-core heptose region induced the susceptibility of Yersinia pestis to the bactericidal action of normal human serum (24). Having determined the functions of all genes involved with the A. hydrophila LPS core biosynthesis pathways and most corresponding single-gene mutants now allows experimental work on the role of the LPS core in the virulence of this bacterium.
Supplementary Material
Acknowledgments
This work was supported by Plan Nacional de I+D and FIS grants (Ministerio de Educación, Ciencia y Deporte and Ministerio de Sanidad, Spain) and by Generalitat de Catalunya. N.J. and R.C. received predoctoral fellowships from Generalitat de Catalunya and Universidad de Barcelona. A.N.K. was supported by the Council on Grants at the President of the Russian Federation for the Support of Young Russian Scientists (project MK-641.2008.4) and B.L. was supported by the Deutscheforschungsgemeinschaft (project Li-448/4-1).
We thank Maite Polo for her technical assistance.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abeyrathne, P. D., C. Daniels, K. K. H. Poon, M. J. Matewish, and J. S. Lam. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 1873002-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, A. G., and D. J. Maskell. 1996. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol. Microbiol. 1937-52. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Austin, B., and C. Adams. 1996. Fish pathogens, p. 197-243. In B. Austin, M. Altwegg, P. J. Gosling, and S. W. Joseph (ed.), The genus Aeromonas. John Wiley and Sons, New York, NY.
- 5.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belunis, C. J., T. Clementz, S. M. Carty, and C. H. R. Raetz. 1995. Inhibition of lipopolysaccharide biosynthesis and cell growth following inactivation of the kdtA gene in Escherichia coli. J. Biol. Chem. 26027646-27652. [DOI] [PubMed] [Google Scholar]
- 7.Canals, R., N. Jiménez, S. Vilches, M. Regué, S. Merino, and J. M. Tomás. 2006. The UDP N-acetylgalactosamine 4-epimerase is essential for mesophilic Aeromonas hydrophila serotype O34 virulence. Infect. Immun. 74537-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caroff, M., and D. Karibian. 2003. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 3382431-2447. [DOI] [PubMed] [Google Scholar]
- 9.Darveau, R. P., and R. E. W. Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galanos, C., O. Lüderitz, and O. Westphal. 1969. A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9245-249. [DOI] [PubMed] [Google Scholar]
- 11.Gronow, S., W. Brabetz, B. Lindner, and H. Brade. 2005. OpsX from Haemophilus influenzae represents a novel type of heptosyltransferase I in lipopolysaccharide biosynthesis. J. Bacteriol. 1876242-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 13.Heinrichs, D. E., J. A. Yethon, and C. Whitfield. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 30221-232. [DOI] [PubMed] [Google Scholar]
- 14.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holst, O. 2002. Chemical structure of the core region of lipopolysaccharides. An update. Trends Glycosci. Glycotechnol. 1487-103. [Google Scholar]
- 16.Izquierdo, L., N. Abitiu, N. Coderch, B. Hita, S. Merino, R. Gavin, J. M. Tomás, and M. Regué. 2002. The inner-core lipopolysaccharide biosynthetic waaE gene: function and genetic distribution among some Enterobacteriaceae. Microbiology 1483485-3496. [DOI] [PubMed] [Google Scholar]
- 17.Izquierdo, L., N. Coderch, N. Piqué, E. Bedini, M. M. Corsaro, S. Merino, S. Fresno, J. M. Tomas, and M. Regué. 2003. The Klebsiella pneumoniae wabG gene: role in biosynthesis of the core lipopolysaccharide and virulence. J. Bacteriol. 1857213-7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Janda, J. M., L. S. Guthertz, R. P. Kokka, and T. Shimada. 1994. Aeromonas species in septicemia: laboratory characteristics and clinical observations. Clin. Infect. Dis. 1977-83. [DOI] [PubMed] [Google Scholar]
- 19.Janda, J. M., S. L. Abbott, S. Khashe, G. H. Kellogg, and T. Shimada. 1996. Further studies on biochemical characteristics and serologic properties of the genus Aeromonas. J. Clin. Microbiol. 341930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27332-344. [DOI] [PubMed] [Google Scholar]
- 21.Kneidinger, B., C. Marolda, M. Graninger, A. Zamyatina, F. McArthur, P. Kosma, M. A. Valvano, and P. Messner. 2002. Biosynthesis pathway of ADP-l-glycero-β-d-manno-heptose in Escherichia coli. J. Bacteriol. 184363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knirel, Y. A., A. S. Shaskov, S. N. Senchenkova, S. Merino, and J. M. Tomás. 2002. Structure of the O-polysaccharide of Aeromonas hydrophila O34: a case of random O-acetylation of 6-deoxy-L-talose. Carbohydr. Res. 3371381-1386. [DOI] [PubMed] [Google Scholar]
- 23.Knirel, Y. A., E. Vinogradov, N. Jimenez, S. Merino, and J. M. Tomás. 2004. Structural studies on the R-type lipopolysaccharide of Aeromonas hydrophila. Carbohydr. Res. 339787-793. [DOI] [PubMed] [Google Scholar]
- 24.Knirel, Y. A., S. V. Dentovskaya, O. V. Bystrova, N. A. Kocharova, S. N. Senchenkova, R. Z. Shaikhutdinova, G. M. Titareva, I. V. Bakhteeva, B. Lindner, G. B. Pier, and A. P. Anisimov. 2007. Relationship of the lipopolysaccharide structure of Yersinia pestis to resistance to antimicrobial factors. Adv. Exp. Med. Biol. 60388-96. [DOI] [PubMed] [Google Scholar]
- 25.Merino, S., S. Camprubí, and J. M. Tomás. 1992. Effect of growth temperature on outer membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infect. Immun. 604343-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milton, D. L., R. O'Toole, P. Hörstedt, and H. Wolf-Watz. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 1781310-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogueras, M. M., S. Merino, A. Aguilar, V. J. Benedi, and J. M. Tomás. 2000. Cloning, sequencing, and role in serum susceptibility of porin II from mesophilic Aeromonas hydrophila. Infect. Immun. 681849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regué, M., N. Climent, N. Abitiu, N. Coderch, S. Merino, L. Izquierdo, M. Altarriba, and J. M. Tomás. 2001. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J. Bacteriol. 1833564-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regué, M., L. Izquierdo, S. Fresno, N. Piqué, M. M. Corsaro, T. Naldi, C. D. Castro, D. Waidelich, S. Merino, and J. M. Tomás. 2005. A second outer-core region in Klebsiella pneumoniae lipopolysaccharide. J. Bacteriol. 187:4198-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubirés, X., F. Saigí, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 1797581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, O. C. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 1888272-8882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vinogradov, E. V., K. Bock, O. Holst, and H. Brade. 1995. The structure of the lipid A-core region of the lipopolysaccharides from Vibrio cholerae O1 smooth strains 569B (Inaba) and rough mutant strain 95R (Ogawa). Eur. J. Biochem. 233152-158. [DOI] [PubMed] [Google Scholar]
- 36.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharide extraction with phenol-water and further application of the procedure. Methods Carbohydr. Chem. 583-89. [Google Scholar]
- 37.Yu, H. B., Y. L. Zhang, Y. L. Lau, F. Yao, S. Vilches, S. Merino, J. M. Tomas, S. P. Howard, and K. Y. Leung. 2005. Identification and characterization of putative virulence genes and gene clusters in Aeromonas hydrophila PPD134/91. Appl. Environ. Microbiol. 714469-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.