Abstract
Accumulation of mutant topoisomerase I cleavage complex can lead to SOS induction and cell death in Escherichia coli. The single-stranded break associated with mutant topoisomerase I cleavage complex is converted to double-stranded break, which then is processed by the RecBCD pathway, followed by association of RecA with the single-stranded DNA.
DNA topoisomerases catalyze the interconversion of different DNA topological forms by coupling DNA strand passage with concerted breaking and rejoining of DNA (5, 43). Escherichia coli DNA topoisomerase I encoded by the topA gene plays an important role in the homeostatic regulation of DNA supercoiling (40, 41). It has an essential function in E. coli for preventing hypernegative supercoiling and R-loop formation (4, 19). Mutants lacking topA function fail to grow at low temperature (20, 37) and readily acquire compensatory mutations (10, 11, 30, 33). Mutants with a topA deletion and compensatory mutations have been shown to be defective in stress response because of their compromised ability to transcribe stress response genes (9, 13, 31, 32, 35, 39, 42). Loss of topA function also results in decreased resistance to extreme low pH (36).
DNA topoisomerases constitute an important class of therapeutic targets of anticancer and antibacterial agents (2, 27, 29). The drugs targeting topoisomerases achieve killing of cancer or bacterial cells not because the drugs inhibit the biological function of topoisomerases but because the drugs stabilize or increase the level of the covalent intermediate formed by topoisomerases with cleaved DNA during the catalytic cycle. Fluoroquinolones are highly potent antibacterial compounds that stabilize the covalent intermediates of DNA gyrase and topoisomerase IV (3, 12, 17). There is at least one type IA topoisomerase found in every bacterium examined thus far that is likely to be required for resolving entanglement of single strands of DNA during replication or recombination (43). Bacterial topoisomerase I could potentially be a useful target for development of novel antibacterial compounds to alleviate the need of new therapeutic drugs. However, since type IA topoisomerase cleaves a single-strand of DNA at a time, it was not clear whether the accumulation of such a cleavage complex would result in lethality for the bacterial cell. The potential of type IA topoisomerases as bactericidal targets was validated when a mutant of Yersinia pestis topoisomerase I, YTOP128, was isolated and characterized (6, 8). This mutant enzyme could cleave DNA and form the covalent complex but failed to religate the cleaved DNA due to a G122S substitution in the TOPRIM domain found among the three mutations identified on the topoisomerase coding sequence of YTOP128. The purified YTOP-G122S enzyme was also active in DNA cleavage but had no religation or relaxation activity (8). Overexpression of either the original YTOP128 mutant or the YTOP-G122S single substitution mutant topoisomerase I in E. coli led to a rapid loss of cell viability (8), with a slightly higher rate of cell killing for YTOP128 (6). These observations support the previous hypothesis that stabilization of covalent complex formed by bacterial type IA topoisomerase can lead to bacterial cell killing (14, 25). The Y. pestis topoisomerase I mutant YTOP128 was isolated originally by its ability to induce the SOS response of E. coli. Events that occur after the formation of the stable topoisomerase I cleavage complex leading to SOS induction, as well as the pathway of cell killing, remain unclear. Such information would shed light on how DNA damage from covalent protein-DNA complexes are repaired in E. coli and would also be useful for future evaluation of potential antibacterial compounds targeting topoisomerase I. The study reported here utilized the TOPRIM topoisomerase I mutant model system of YTOP128 and YTOP-G122S to examine the cellular response to stabilized topoisomerase I complex in E. coli.
Induction of SOS response by expression of YTOP128 and YTOP-G122S.
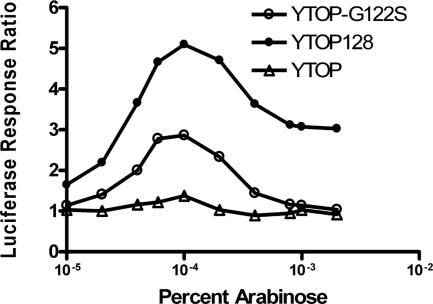
The dinD1::luxCDABE on plasmid pDinlux (7) was utilized as a reporter of SOS induction. Expression of YTOP128 and YTOP-G122S from the BAD promoter on pAYTOP128 and pAYTOP-G122S (7) in E. coli strain BW27784 (15) with increasing concentrations of arabinose resulted in increase of luciferase signal (Fig. 1). There was no significant SOS induction from the expression of wild-type YTOP. The luciferase signal reached a maximum value at ca. 0.00006 to 0.0002% arabinose for both YTOP128 and YTOP-G122S. A further increase in arabinose concentration added to the culture resulted in drop of luciferase signal. This is probably due to the loss of cell viability resulting from the accumulation of high levels of topoisomerase I cleavage complex, since DNA religation is inhibited as a result of the G122S mutation (8). The presence of the M326V mutation in YTOP128 enhances DNA cleavage, with a 10- to 40-fold higher rate of cell killing (6) and also a higher luciferase activity from SOS induction (Fig. 1).
FIG. 1.
Luciferase signal as reporter of SOS induction by mutant topoisomerase I cleavage complex. BW27784 transformed with luciferase reporter plasmid pDinlux along with either plasmid pAYOP (▵), plasmid pAYTOP128 (•), or plasmid pAYTOP-G122S (○) was grown to exponential phase in LB medium with ampicillin and chloramphenicol at 37°C until the A600 reached 0.4. The culture was dispensed in 50-μl aliquots into white 96-well microtiter plates. Equal volume of LB medium containing 0.00001 to 0.004% arabinose was added for induction of recombinant Y. pestis topoisomerase I. The light production from the induced luciferase was measured on a Perkin-Elmer 7000 Plus BioAssay reader at 37°C in 10 min cycles with 30 s of shaking duration before each measurement. The luciferase response ratio was measured as the ratio of luciferase signal from the treated cultures versus the same culture that has not been treated with arabinose at 260 min after the addition of arabinose.
Effect of recA-null mutation on SOS induction and cell killing by topoisomerase I cleavage complex.
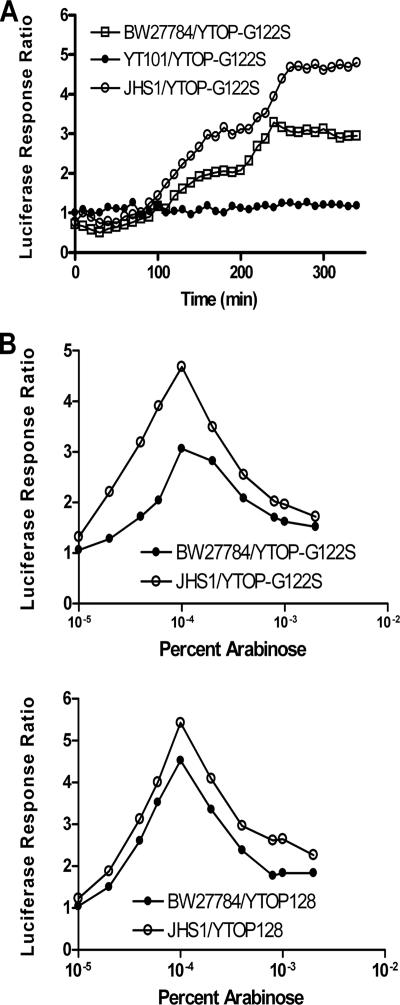
We examined the effect of a complete loss of recA function on the cellular response to accumulated topoisomerase I cleavage complex. The constructions of the mutant derivatives of BW27784 are listed in Table S1 in the supplemental material. As shown in Fig. 2A, the luciferase signal from the dinD1 promoter activity upon arabinose induction of YTOP-G122S was abolished in strain YT101 (recA::Tn5). The loss of SOS induction in YT101 was accompanied by a several-hundredfold decrease in cell viability measured 2 h after the addition of arabinose compared to the BW27784 parent (Table 1). These results from strain YT101 demonstrated not only that RecA function is involved in SOS induction after the accumulation of the topoisomerase I cleavage complex but also that RecA plays a role in protecting the bacterial cell from the killing effect of the topoisomerase I cleavage complex accumulation.
FIG. 2.
Comparison of induction of dinD1::luxCDABE in strains with different recA genotypes after treatment with arabinose for the expression of recombinant mutant Y. pestis topoisomerase I. (A) The luciferase response ratio was measured in BW27784 (recA+), YT101 (recA::Tn5), or JHS1 (recA718) as the ratio of luciferase signal from dinD1::luxCDABE in cultures induced with 0.0001% arabinose for the expression of YTOP-G122S versus luciferase signal from the same culture not treated with arabinose. (B) Luciferase response signal measured at 260 min after the addition of different concentrations of arabinose in BW27784 or JHS1 transformed with pAYTOP-G122S or pAYTOP128.
TABLE 1.
Effect of recA mutations on relative cell viability following induction of wild-type or mutant Y. pestis topoisomerase I
| Strain | Avg relative viability ± SDa
|
|||
|---|---|---|---|---|
| pAYTOP with:
|
pAYTOP-G122S with:
|
|||
| 0.00006% arabinose | 0.0002% arabinose | 0.00006% arabinose | 0.0002% arabinose | |
| BW27784 | 0.87 ± 0.30 | 0.45 ± 0.20 | 0.065 ± 0.007 | 0.020 ± 0.003 |
| YT101 | 1.05 ± 0.15 | 0.52 ± 0.19 | 3.9 × 10−4 ± 1.5 × 10−4 | 6.0 × 10−5 ± 3.3 × 10−5 |
| JHS1 | 0.71 ± 0.40 | 0.19 ± 0.07 | 9.2 × 10−3 ± 5.3 × 10−3 | 1.1 × 10−4 ± 3.4 × 10−5 |
The relative viability was measured as the ratio of viable counts determined 2 h after the addition of 0.00006 or 0.0002% arabinose versus the viable counts from control cultures not treated with arabinose. The results represent the average from three experiments.
SOS induction is retained in the recA718 mutant strain, but cell survival is compromised.
To separate the SOS induction ability of RecA protein from its other functions in DNA repair, the recA718 mutation was introduced into BW27784. Like wild-type recA, this allele requires DNA damage to become activated for SOS induction (21, 23). Luciferase measurement from dinD1::luxCDABE fusion after induction of YTOP-G122S with arabinose confirmed that strain JHS1 carrying the recA718 allele responded to the topoisomerase I-mediated DNA damage with SOS induction (Fig. 2A). The luciferase response ratio was found to be slightly higher after the induction of both YTOP-G122S and YTOP128 in JHS1 than in BW27784 for arabinose concentrations between 0.00001 and 0.002% (Fig. 2B). Compared to recA+ strains, strains with recA718 mutation showed moderate UV sensitivity (45, 46), and the recA718 mutation was lethal when combined with certain polA mutations (44) due to the effect of the recA718 mutation on homologous recombination. It was hypothesized that RecA718 may perform certain recombinational repair processes less efficiently than wild-type RecA (44). JHS1 also showed a greatly decreased survival rate after the induction of YTOP-G122S compared to BW27784 (Table 1). These results from JHS1 showed that RecA homologous recombination function is directly involved in the repair of the topoisomerase I-mediated DNA lesion and that the SOS induction function of RecA is by itself insufficient for evading lethality due to the topoisomerase I-mediated DNA cleavage.
Extensive chromosomal fragmentation from the topoisomerase I-mediated DNA lesion.
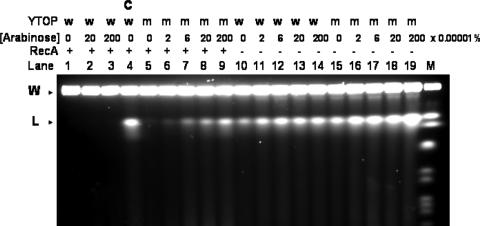
Chromosomal fragmentation occurs in E. coli in many situations when DNA lesions are formed and when recA is inactivated by mutation (16). If chromosomal fragmentation is increased significantly after trapping of the topoisomerase I cleavage complex, it would account for the higher rate of cell killing in YT101 when mutant topoisomerase I was induced. Pulsed-field gel electrophoresis was carried out under conditions such that the intact chromosomes stay in the wells, while the linear chromosomes enter the gel and the chromosomal fragments form a smear (16, 24, 38). None of the DNA entered the gel after induction of wild-type YTOP in BW27784 for 30 min with 0.0002 and 0.002% arabinose (Fig. 3, lanes 2 to 3). Linear chromosome and fragmented DNA can be observed after treatment with 0.5 mg of ciprofloxacin/liter for 30 min (lane 4). When mutant YTOP128 was induced in BW27784 with 0.00002 to 0.002% arabinose (lanes 6 to 9), chromosomal fragmentation could be observed after 30 min of incubation. There is some level of background DNA fragmentation in YT101 due to the recA-null mutation even with no arabinose added (lanes 10). When wild-type YTOP was induced in YT101 (lanes 11 to 14), additional chromosomal fragmentation over the background level could be observed, suggesting that, with the RecA function missing, overexpression of wild-type recombinant Y. pestis topoisomerase I could also result in increased DNA breaks from the large number of topoisomerase I cleavage events in the bacterial DNA genome. However, overexpression of wild-type recombinant Y. pestis topoisomerase I in BW27784 with the recA+ genotype did not produce any DNA breaks (lanes 2 to 3). The chromosomal fragmentation in YT101 after the induction of YTOP128 by arabinose (lanes 16 to 19) could be readily observed as a smear of DNA entering the gel in addition to the linear chromosomal DNA, indicating the extensive accumulation of DNA breaks resulting from the cleavage complex formed by the mutant topoisomerase I. Western blot analysis of the total cellular proteins at 30 min after addition of arabinose showed that the expression levels of recombinant topoisomerase I with increasing arabinose concentrations were similar for wild-type YTOP and YTOP128 (see Fig. S1 in the supplemental material).
FIG. 3.
Chromosomal fragmentation measured by pulsed-field gel electrophoresis. Exponential-phase cultures were induced with arabinose for the expression of wild-type and mutant recombinant Y. pestis topoisomerase I. After incubation with shaking at 37°C for 30 min, cells were collected by centrifugation and processed with the Bio-Rad CHEF bacterial genomic DNA plug kit. The agarose plugs containing the genomic DNA were electrophoresed in a 1% agarose gel with 0.5× Tris-borate-EDTA buffer with the Bio-Rad CHEF-DRII pulsed-field electrophoresis system (14°C, 6 V/cm, 21 h, linear ramping from 50 to 92 s), followed by staining with ethidium bromide. Analysis was carried out for BW27784 (lanes 1 to 9) or YT101 (recA::Tn5, lanes 10 to 19) cells transformed with pAYTOP (w, lanes 1 to 4 and lanes 10 to 14) or pAYTOP128 (m, lanes 5 to 9 and lanes 15 to 19). Chromosomal DNA gel plugs were prepared after 30 min of no treatment (lanes 1, 10, and 15), treatment with 0.5 mg ciprofloxacin/liter (C, lane 4), or arabinose at the indicated concentrations. W, gel wells; L, linear chromosome; M, Saccharomyces cerevisiae chromosome standards (225 to 2,200 kb).
Requirement of RecBCD instead of the RecFOR complex for SOS induction and DNA repair after the accumulation of topoisomerase I-mediated DNA lesions.
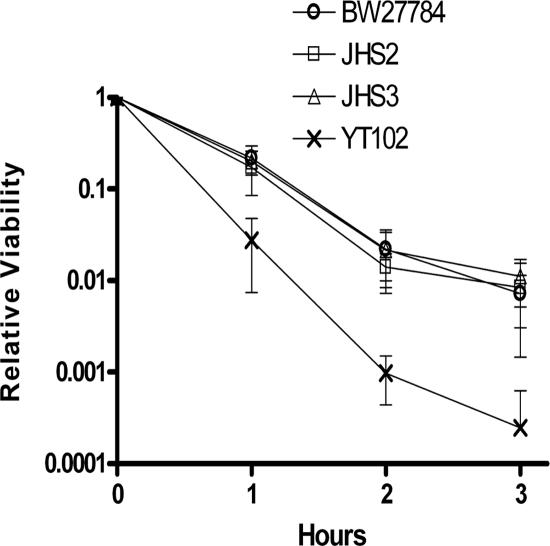
In E. coli, the RecA homologous recombination can be initiated after DNA damage via either the RecBCD pathway or the RecFOR pathway (34). The RecBCD complex is involved in processing of double-stranded DNA breaks before RecA catalyzed repair can take place (1). The RecFOR complex is mainly involved in single-strand gap repair in E. coli (34). After the treatment of E. coli with drug compounds that trap the covalent cleavage complexes formed by DNA gyrase, RecBC functions are required for SOS induction (26). However, DNA gyrase is a type IIA topoisomerase that cleaves a double strand of DNA at a time, while cleavage by the topoisomerase I would form a single-strand break on DNA. It is possible that repair of the topoisomerase I-mediated DNA break may involve the RecFOR gap repair pathway. We therefore measured the effects of recB, recF, and recR mutations in the BW27784 genetic background on SOS induction by the mutant YTOP cleavage complex. Luciferase measurement from dinD1::luxCDABE (Table 2) showed that SOS induction could not be observed in the presence of the recB mutation. The RecBCD pathway of double-stranded break repair thus appears to be required for generating the single-stranded DNA that becomes the substrate of RecA filament formation. In contrast, recF or recR mutation had little effect on SOS induction by the accumulated mutant topoisomerase I cleavage complex (Table 2), suggesting that the RecFOR gap repair pathways is not involved in the response. These conclusions from the SOS induction measurements were further supported by comparison of the viability of the recB mutant derivatives of BW27784 (YT102) versus the recF (JHS2) and recR (JHS3) mutants after induction of mutant YTOP-G122S (Fig. 4).
TABLE 2.
Effect of recB, recF, and recR mutations on luciferase response ratio from SOS induction by mutant topoisomerase I cleavage complex
| Strain | Luciferase response ratioa
|
|
|---|---|---|
| YTOP128 | YTOP-G122S | |
| BW27784 | 4.7 | 3.5 |
| YT102 (recB) | 0.91 | 0.96 |
| JHS2 (recF) | 4.1 | 3.4 |
| JHS3 (recR) | 4.4 | 3.9 |
Luciferase response ratio was measured as the ratio of luminescence measured at 260 min after the addition of 0.00006% arabinose versus the luminescence from control cultures not treated with arabinose.
FIG. 4.
Effect of recB, recF, or recR mutations on cell viability after induction of mutant Y. pestis topoisomerase I. Arabinose (0.002%) was added to the exponential-phase cultures of BW27784, YT102, JHS2, and JHS3 transformed with pAYTOP-G122S. Viable cell counts of arabinose-treated cultures determined after different periods of incubation at 37°C were divided by viable cell counts from untreated cultures to determine the relative viability. The results represent the averages and standard deviations from three experiments.
Similarity in cell killing pathway of type IA and type IIA topoisomerases.
The formation of double-stranded breaks that are substrates of RecBCD is similar to the cell killing pathway of fluoroquinolone action (18, 26, 28). The conversion of the single-stranded break associated with topoisomerase I to a double-stranded break probably accounts for the rapid cell killing by the accumulated topoisomerase I-mediated DNA lesion due to the TOPRIM Gly-to-Ser topoisomerase I mutation (8).
Supplementary Material
Acknowledgments
This study was supported by Public Health Service grant R01 AI 069313 from the National Institutes of Health.
We thank R. Woodgate and S. Sandler for kindly providing bacterial strains and T. Annamalai for helpful comments on the manuscript.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 9077-86. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, V. E., and N. Osheroff. 2001. Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr. Pharm. Des. 7337-353. [DOI] [PubMed] [Google Scholar]
- 3.Aubry, A., X. S. Pan, L. M. Fisher, V. Jarlier, and E. Cambau. 2004. Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob. Agents Chemother. 481281-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baaklini, I., C. Hraiky, F. Rallu, Y. C. Tse-Dinh, and M. Drolet. 2004. RNase HI overproduction is required for efficient full-length RNA synthesis in the absence of topoisomerase I in Escherichia coli. Mol. Microbiol. 54198-211. [DOI] [PubMed] [Google Scholar]
- 5.Champoux, J. J. 2001. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70369-413. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, B., Sorokin, E., and Y. C. Tse-Dinh. 2008. Mutation adjacent to the active site tyrosine can enhance DNA cleavage and cell killing by the TOPRIM Gly to Ser mutant of bacterial topoisomerase I. Nucleic Acids Res. 361017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, B., I. Liu, and Y. C. Tse-Dinh. 2007. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J. Antimicrob. Chemother. 59640-645. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, B., S. Shukla, S. Vasunilashorn, S. Mukhopadhyay, and Y. C. Tse-Dinh. 2005. Bacterial cell killing mediated by topoisomerase I DNA cleavage activity. J. Biol. Chem. 28038489-38495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, B., S. Rui, C. Ji, V. W. Gong, T. K. Van Dyk, M. Drolet, and Y. C. Tse-Dinh. 2003. RNase H overproduction allows the expression of stress-induced genes in the absence of topoisomerase I. FEMS Microbiol. Lett. 221237-242. [DOI] [PubMed] [Google Scholar]
- 10.DiNardo, S., K. A. Voelkel, R. Sternglanz, A. E. Reynolds, and A. Wright. 1982. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 3143-51. [DOI] [PubMed] [Google Scholar]
- 11.Dorman, C. J., A. S. Lynch, N. N. Bhriain, and C. F. Higgins. 1989. DNA supercoiling in Escherichia coli: topA mutations can be suppressed by DNA amplifications involving the tolC locus. Mol. Microbiol. 3531-540. [DOI] [PubMed] [Google Scholar]
- 12.Drlica, K. 1999. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2504-508. [DOI] [PubMed] [Google Scholar]
- 13.Drolet, M. 2006. Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol. Microbiol. 59723-730. [DOI] [PubMed] [Google Scholar]
- 14.Fernandes, P. B., R. Menzel, D. J. Hardy, Y. C. Tse-Dinh, A. Warren, and D. A. Elsemore. 1999. Microbial resistance: novel screens for a contemporary problem. Med. Res. Rev. 19559-568. [DOI] [PubMed] [Google Scholar]
- 15.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. 2001. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 1473241-3247. [DOI] [PubMed] [Google Scholar]
- 16.Kouzminova, E. A., E. Rotman, L. Macomber, J. Zhang, and A. Kuzminov. 2004. RecA-dependent mutants in Escherichia coli reveal strategies to avoid chromosomal fragmentation. Proc. Natl. Acad. Sci. USA 10116262-16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine, C., H. Hiasa, and K. J. Marians. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim. Biophys. Acta 140029-43. [DOI] [PubMed] [Google Scholar]
- 18.Malik, M., X. Zhao, and K. Drlica. 2006. Lethal fragmentation of bacterial chromosomes mediated by DNA gyrase and quinolones. Mol. Microbiol. 61810-825. [DOI] [PubMed] [Google Scholar]
- 19.Masse, E., and M. Drolet. 1999. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J. Biol. Chem. 27416654-16658. [DOI] [PubMed] [Google Scholar]
- 20.Masse, E., and M. Drolet. 1999. R-loop-dependent hypernegative supercoiling in Escherichia coli topA mutants preferentially occurs at low temperatures and correlates with growth inhibition. J. Mol. Biol. 294321-332. [DOI] [PubMed] [Google Scholar]
- 21.McCall, J. O., E. M. Witkin, T. Kogoma, and V. Roegner-Maniscalco. 1987. Constitutive expression of the SOS response in recA718 mutants of Escherichia coli requires amplification of RecA718 protein. J. Bacteriol. 169728-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reference deleted.
- 23.McLenigan, M. P., O. I. Kulaeva, D. G. Ennis, A. S. Levine, and R. Woodgate. 1999. The bacteriophage P1 HumD protein is a functional homolog of the prokaryotic UmuD′-like proteins and facilitates SOS mutagenesis in Escherichia coli. J. Bacteriol. 1817005-7013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaraja, V., D. Sikder, and P. Jain. 2002. DNA topoisomerase I from mycobacteria: a potential drug target. Curr. Pharm. Des. 81995-2007. [DOI] [PubMed] [Google Scholar]
- 26.Newmark, K. G., E. K. O'Reilly, J. R. Pohlhaus, and K. N. Kreuzer. 2005. Genetic analysis of the requirements for SOS induction by nalidixic acid in Escherichia coli. Gene 35669-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nitiss, J. L. 2002. DNA topoisomerases in cancer chemotherapy: using enzymes to generate selective DNA damage. Curr. Opin. Investig. Drugs 31512-1516. [PubMed] [Google Scholar]
- 28.Pohlhaus, J. R., and K. N. Kreuzer. 2005. Norfloxacin-induced DNA gyrase cleavage complexes block Escherichia coli replication forks, causing double-stranded breaks in vivo. Mol. Microbiol. 561416-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pommier, Y. 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6789-802. [DOI] [PubMed] [Google Scholar]
- 30.Pruss, G. J., S. H. Manes, and K. Drlica. 1982. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell 3135-42. [DOI] [PubMed] [Google Scholar]
- 31.Qi, H., R. Menzel, and Y. C. Tse-Dinh. 1999. Increased thermosensitivity associated with topoisomerase I deletion and promoter mutations in Escherichia coli. FEMS Microbiol. Lett. 178141-146. [DOI] [PubMed] [Google Scholar]
- 32.Qi, H., R. Menzel, and Y. C. Tse-Dinh. 1996. Effect of the deletion of the sigma 32-dependent promoter (P1) of the Escherichia coli topoisomerase I gene on thermotolerance. Mol. Microbiol. 21703-711. [DOI] [PubMed] [Google Scholar]
- 33.Raji, A., D. J. Zabel, C. S. Laufer, and R. E. Depew. 1985. Genetic analysis of mutations that compensate for loss of Escherichia coli DNA topoisomerase I. J. Bacteriol. 1621173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocha, E. P., E. Cornet, and B. Michel. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rui, S., and Y. C. Tse-Dinh. 2003. Topoisomerase function during bacterial responses to environmental challenge. Front. Biosci. 8d256-d263. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, N., J. Feng, X. Liu, D. Chaudhuri, J. W. Foster, M. Drolet, and Y. C. Tse-Dinh. 2005. Loss of topoisomerase I function affects the RpoS-dependent and GAD systems of acid resistance in Escherichia coli. Microbiology 1512783-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stupina, V. A., and J. C. Wang. 2005. Viability of Escherichia coli topA mutants lacking DNA topoisomerase I. J. Biol. Chem. 280355-360. [DOI] [PubMed] [Google Scholar]
- 38.Thoms, B., and W. Wackernagel. 1998. Interaction of RecBCD enzyme with DNA at double-strand breaks produced in UV-irradiated Escherichia coli: requirement for DNA end processing. J. Bacteriol. 1805639-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tse-Dinh, Y. C. 2000. Increased sensitivity to oxidative challenges associated with topA deletion in Escherichia coli. J. Bacteriol. 182829-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tse-Dinh, Y. C. 1985. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 134751-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tse-Dinh, Y. C., and R. K. Beran. 1988. Multiple promoters for transcription of the Escherichia coli DNA topoisomerase I gene and their regulation by DNA supercoiling. J. Mol. Biol. 202735-742. [DOI] [PubMed] [Google Scholar]
- 42.Tse-Dinh, Y. C., H. Qi, and R. Menzel. 1997. DNA supercoiling and bacterial adaptation: thermotolerance and thermoresistance. Trends Microbiol. 5323-326. [DOI] [PubMed] [Google Scholar]
- 43.Wang, J. C. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell. Biol. 3430-440. [DOI] [PubMed] [Google Scholar]
- 44.Witkin, E. M., and V. Roegner-Maniscalco. 1992. Overproduction of DnaE protein (alpha subunit of DNA polymerase III) restores viability in a conditionally inviable Escherichia coli strain deficient in DNA polymerase I. J. Bacteriol. 1744166-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Witkin, E. M., V. Roegner-Maniscalco, J. B. Sweasy, and J. O. McCall. 1987. Recovery from ultraviolet light-induced inhibition of DNA synthesis requires umuDC gene products in recA718 mutant strains but not in recA+ strains of Escherichia coli. Proc. Natl. Acad. Sci. USA 846805-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witkin, E. M., J. O. McCall, M. R. Volkert, and I. E. Wermundsen. 1982. Constitutive expression of SOS functions and modulation of mutagenesis resulting from resolution of genetic instability at or near the recA locus of Escherichia coli. Mol. Gen. Genet. 18543-50. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.