Abstract
The peptidyl transferase center, present in domain V of 23S rRNA of eubacteria and large rRNA of plants and animals, can act as a general protein folding modulator. Here we show that a few specific nucleotides in Escherichia coli domain V RNA bind to unfolded proteins and, as shown previously, bring the trapped proteins to a folding-competent state before releasing them. These nucleotides are the same for the proteins studied so far: bovine carbonic anhydrase, lactate dehydrogenase, malate dehydrogenase, and chicken egg white lysozyme. The amino acids that interact with these nucleotides are also found to be specific in the two cases tested: bovine carbonic anhydrase and lysozyme. They are either neutral or positively charged and are present in random coils on the surface of the crystal structure of both the proteins. In fact, two of these amino acid-nucleotide pairs are identical in the two cases. How these features might help the process of protein folding is discussed.
Ribosomes from various sources have shown protein-folding activity in vitro (1, 13, 14, 23). This activity was tested on more than a dozen proteins with various tertiary structures and organizations (1, 7, 14, 20). The activity was found later to be present in the large ribosomal subunit, and on stripping ribosomes of their proteins, it could be assigned to the peptidyl transferase center (PTC) in the larger RNA of the large subunit (8, 11, 30, 35). In Escherichia coli and in other eubacteria in general, the PTC is in domain V (U2016 to G2625 of E. coli) of 23S rRNA.
If the same RNA can fold a large number of proteins which are structurally unrelated, the question of specificity arises in the interaction of the unfolded proteins with the RNA. Here we have deciphered the RNA-protein interactions at the nucleotide-amino acid level.
To locate the interacting nucleotides of domain V, we used primer extension analysis and determined the positions where the extensions were blocked. We found that the same set of nucleotides interacts with four different proteins tested: bovine carbonic anhydrase (BCA), lactate dehydrogenase (LDH), malate dehydrogenase (MDH), and chicken egg white lysozyme. Using matrix-assisted laser desorption ionization-time of flight/time of flight (MALDI-TOF/TOF), we also tested whether the RNA interacted with specific amino acids. In the two proteins tested, BCA and lysozyme, the amino acids involved were specific for each protein and could even be identical between the two proteins. The disposition of the amino acids on the three-dimensional structure shared some common features. The RNA-protein interactions thus appear to have significant specificity.
MATERIALS AND METHODS
In vitro synthesis of domain V of E. coli 23S rRNA.
The rRNA gene fragment containing domain V was cloned in vector pTZ57R/T (Fermentas) under the control of a T7 promoter. The presence of the domain in selected clones was verified by DNA sequencing. An EcoRI restriction site, just downstream of the domain, was used to linearize the plasmid, and RNA was synthesized using T7 RNA polymerase. Similar cloning and synthesis were done for domain II (G559 to G1283) and domain IV (C1670 to G2029) of E. coli 23S rRNA.
Unfolding and refolding of proteins.
BCA, LDH, MDH, lysozyme, and human carbonic anhydrase I (HCA-I) were denatured with guanidine hydrochloride (7, 11, 31, 35), and the loss of their secondary structures was verified by circular dichroism spectra (16). For all enzymes except LDH, 6 M guanidine hydrochloride was used for denaturation. For LDH, 1 M denaturant was used. The denatured enzymes were diluted 100 times (final concentration, 300 nM) in the refolding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 10 mM magnesium acetate) in the absence or in the presence of 23S or domain V rRNA and were incubated for 30 min at 25°C to allow folding. The molar concentration of RNA was twice that of the enzymes. The residual amount of guanidine hydrochloride had no effect on the activities of the enzymes. The activities of refolded proteins were determined, following published protocols (7, 11, 31, 35), and expressed as a percentage of activity of the same amount of native protein. For lysozyme the activity assay was not done, but cross-linking with domain V for primer extension (see below) was done with it.
UV cross-linking of domain V-refolding protein complexes.
Three hundred microliters of sample, containing 125 nM domain V RNA and 600 nM unfolded protein (to ensure that all RNA molecules were bound to proteins), was irradiated on a glass dish in ice with 254-nm UV (GS Gene Linker; Bio-Rad) at a distance of 6 cm for 2 min. The irradiated samples were precipitated by salt-ethanol and washed with 70% ethanol.
Primer extension on cross-linked domain V-protein complexes.
Primer BG32 (5′ACCCCGGAATTCGCGCCCACGGCAGATAGG3′) was annealed to cross-linked domain V-protein complexes (about 3 to 5 μg). Annealed primers were labeled with [α-32P]dCTP, following the 3-deoxynucleoside triphosphate method, at 55°C using ThermoScript reverse transcriptase (Invitrogen). Labeled primers were extended at 58°C after the addition of all four deoxynucleoside triphosphates in excess by the same enzyme for about 45 min. The products were precipitated, washed with 70% ethanol, and analyzed on a 6.5% polyacrylamide gel in 8 M urea next to a sequencing ladder of domain V rRNA genes. It was obtained using the same primer by Thermo Sequenase DNA polymerase (Thermo Sequenase cycle sequencing kit; USB).
Sample preparation for MS.
The mass spectrometry (MS) samples were prepared as described previously (38, 39). UV-irradiated samples were precipitated, air-dried, and dissolved in 8 M urea, 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, and 1 mM dithiothreitol, heated for 10 min at 70°C, and cooled to room temperature. The sample was diluted 10-fold with 50 mM Tris-HCl (pH 8.0) and then digested with trypsin (Gold MS grade; Promega) with an enzyme/substrate ratio of 1:25 for 16 h at 37°C. The samples were precipitated and dissolved in 50 mM Tris-HCl (pH 8.0) and 2 mM EDTA. They were then digested with RNase A and RNase T1 for 2 h at 50°C, followed by trypsin for 16 h at 37°C. Digestion was stopped by direct injection of 500 μl sample onto a μRPC_C18_ST_4.6/100 column (GE Healthcare) mounted in an AKTAbasic 100 system and run using the Unicorn software. For reverse-phase chromatography, the solvents were as follows: solution A, 0.1% (vol/vol) trifluoroacetic acid (TFA) in double-distilled H2O; solution B, 0.085% (vol/vol) TFA in acetonitrile. The column was equilibrated with a 5× column volume (CV) of solution B, and the following gradients were applied in steps with a flow rate of 0.1 ml/min: (i) isocratic elution of the injection peak at the rate of 5% solution B for 2× CV, (ii) 5% to 100% solution B for 20× CV, and (iii) isocratic wash with 10% B until the baseline level was reached. Five hundred microliters of fractions were collected, and the presence of cross-linked peptide-RNA was checked by absorbance at 250 nm. Peak fractions were pooled, vacuum dried, and then mixed with the appropriate matrix for MS and tandem MS (MS/MS) analysis.
MS.
MS was carried out in an Applied Biosystems AB4700 MALDI-TOF/TOF mass spectrometer. Each 0.5-μl sample was mixed on a stainless-steel plate with an equal volume of either 10 mg/ml 2,5-dihydroxy benzoic acid or 5 mg/ml α-cyano-4-hydroxycinnamic acid (recrystallized and dissolved in 50% acetonitrile-0.1% TFA) matrix solutions. It was then dried at room temperature and subjected to MS and MS/MS. PMF were obtained in positive reflector mode. The instrument was operated in the delayed extraction mode with a delay time of 200 ns. Spectra were obtained by accumulation of 2,500 and 6,000 consecutive laser shots, respectively, in MS and MS/MS mode, and the laser intensities used were in the range of 4,000 to 5,000. Close external calibration for MS was performed using 4700 Cal mix (Applied Biosystems). Peak harvesting was carried out using the 4000 series Explorer (Applied Biosystems) software. Collected spectra were processed using Data Explorer software V.4.6 for advanced baseline correction (peak width, 32; flexibility, 0.5; degree, 0.1), noise reduction (correlation factor, 0.8), and deisotoping. Processed spectra were checked for the presence of tryptic peptides and their possible modifications for the representative proteins using the MassSorter v.1.05.04 software program (3). The search parameters included 1 missed cleavage and an error tolerance of ±50 ppm for PMF or ±0.2 Da for the MS/MS ion search.
RESULTS
Refolding of unfolded proteins.
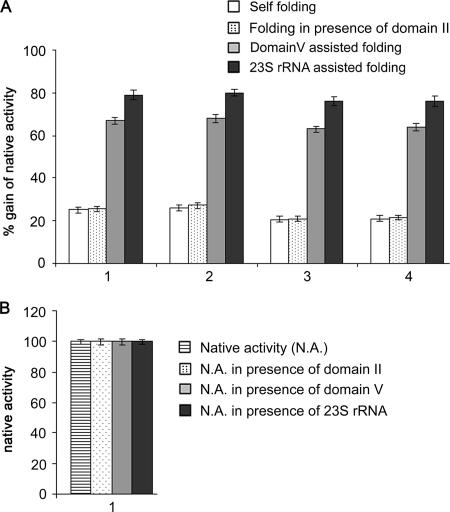
BCA, LDH, MDH, and HCA-I were unfolded with guanidine hydrochloride and diluted 100-fold in the refolding buffer in the absence or in the presence of E. coli 23S rRNA or its domain V or domain II region. The activities of the proteins were measured and expressed in terms of the activities of the same amounts of native proteins (Fig. 1A). The activities due to self-folding were around 20 to 25% of the native activity, while in the case of domain V- and 23S rRNA-assisted folding, the activities increased to around 65 to 70% and 75 to 80%, respectively. The RNA molecules were used at twice the molar concentration of the enzymes. This was necessary to get a better yield of folding. This folding was specific for domain V of 23S rRNA. 16S rRNA, tRNA, and other regions of 23S rRNA (as shown in the case of domain II) did not help to fold the proteins (7, 13). The activities of native proteins did not change when they were incubated with the ribosomal components (shown for BCA only) (Fig. 1B). The folding activity of the RNA could be destroyed by RNase A (7). These results extend our earlier finding of general refolding activity of domain V.
FIG. 1.
Folding of proteins in the presence of rRNA components. (A) Unfolded proteins are folded in the absence and presence of 23S rRNA components. 1, 2, 3, and 4 represent the folding of BCA, HCA-I, MDH, and LDH, respectively. Bar graphs represent the mean folding values (± standard deviations) from three independent experiments. (B) Bar graphs showing the activity of native BCA after incubation without or with different components of 23S rRNA. Bar graphs represent the mean folding values (± standard deviations) from three independent experiments.
Identification of sites on domain V RNA where proteins bind during refolding.
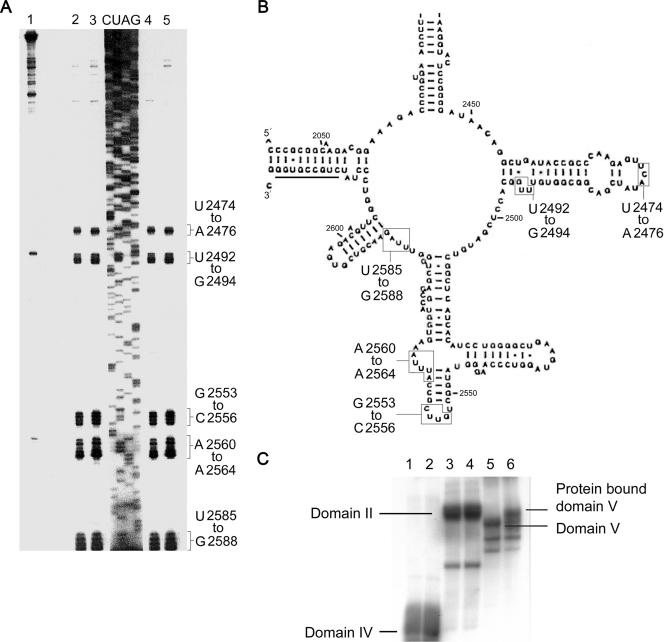
To find the nucleotides which interact with proteins during the refolding process, primer extension studies were done with UV-cross-linked domain V-protein complexes. A set of five distinct clusters of strong reverse transcriptase stops was identified (Fig. 2A). The bases were mostly in the central loop region of PTC and were found to be highly conserved through evolution in prokaryotes (Fig. 2B). The bases that comprise the clusters are as follows: (i) U2474, C2475, and A2476; (ii) U2492, U2493, and G2494; (iii) G2553, U2554, U2555, and C2556; (iv) A2560, U2561, U2562, U2563, and A2564; and (v) U2585, U2586, A2587, and G2588. Among these, the positions G2553, U2555, U2492, and U2493 are known to be required for tRNA binding and translational fidelity (19, 28, 29). The stops were identical in positions and relative intensities for BCA, LDH, MDH, and lysozyme and were absent when RNA was irradiated in the presence of native proteins (data not shown) or in the absence of any protein (Fig. 2A, lane 1).
FIG. 2.
Primer extension analysis on domain V RNA cross-linked to proteins. (A) Primer extension analysis on domain V RNA after UV irradiation in the absence (lane 1) or in the presence (lanes 2 to 5) of protein. The proteins used were BCA, lysozyme, MDH, and LDH in lanes 2 to 5, respectively. (B) Secondary structure of the central loop of E. coli domain V rRNA. The boxes include the nucleotides that are identified to be cross-linked with proteins. The position of the cDNA primer used for primer extension analysis is indicated by the solid line at the 3′ end of domain V. (C) Electrophoretic mobility shift assay showing RNAs UV irradiated without or with BCA. Lanes 1, 3, and 5 represent domain IV, domain II, and domain V of 23S rRNA UV irradiated without BCA, respectively. Lanes 2, 4, and 6 represent the same RNA molecules UV irradiated in the presence of unfolded BCA, respectively. Note that only domain V RNA shows a mobility shift in the presence of unfolded protein (lane 6).
In a separate experiment, we subjected the [α-32P]UTP-labeled domain V, II, and IV regions of 23S rRNA synthesized in vitro to UV cross-linking in the absence and in the presence of unfolded BCA. The UV-irradiated samples were electrophoresed through a 5% native polyacrylamide gel (Fig. 2C). In the presence of the protein, the mobility of only domain V was shifted. This shows that unfolded proteins cross-link specifically with domain V RNA. A few shorter RNA species appeared in the lane of domain V RNA (Fig. 2C, lanes 5 and 6) due to premature termination of transcription, which is normal in cases of transcripts with profuse secondary structures. However, the electrophoretic mobility of these short transcripts was not shifted, apparently because they could not bind the protein.
Strategy for identification of amino acids that interact with domain V by MALDI-TOF MS.
We used the method improvised by the group of Urlaub and Luhrmann to identify oligopeptide stretches that cross-link to domain V RNA during the refolding reaction (24, 34, 37-39). Subsequently, the cross-linked amino acid residues of the oligopeptides were identified. BCA and lysozyme were chosen for these studies since they function as monomers.
UV-irradiated samples were digested with trypsin and then treated with an excess of RNase A and RNase T1 to hydrolyze the RNA, so that the peptides remain attached to a single nucleotide or short oligonucleotides only. The sample was fractionated, and the peak fractions at 250 nm were analyzed for peptides or peptides attached to a nucleotide(s). An accurate analysis of the molecular mass of such cross-linked heteroconjugates, by comparison with the theoretical masses of all possible tryptic fragments of the protein, should provide the composition of the cross-linked peptides. Peptide fragments generated from contaminants such as trypsin, keratin, RNase, etc., were excluded from the analysis.
Accurate mass determination of cross-linked peptide by high-resolution MALDI-TOF spectra.
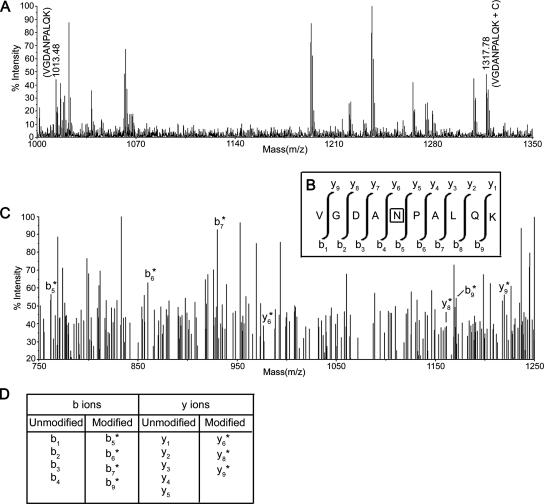
To identify the modified peptide stretches (due to the attached nucleotide), all experimental data were compared with the theoretical data in the presence of custom modifications for our study (nucleotides of RNA and their fragments, such as adenosine [A], A-phosphate, A-ribose, ribose only, etc.) using the MassSorter software program (3). A few modifications and the molecular weights are given in Table S1 in the supplemental material. For both BCA and lysozyme, analyses were performed separately through MassSorter. Five modified stretches were found in both cases. For example, the oligopeptide stretch 148 VGDANPALQK 157 (m/z 1012.48) in the tryptic pool of BCA remains attached with one cytosine (molecular weight, 305) residue, giving rise to a high-intensity spectrum at m/z 1317.78 (Fig. 3A). The interacting stretches of BCA and lysozyme obtained from MassSorter analysis are shown in Table S2 in the supplemental material. MS results clearly showed specific stretches of BCA and lysozyme interacting with domain V during folding.
FIG. 3.
MS and MS/MS analysis to show that the peptide stretch 148 VGDANPALQK 157 of BCA remains cross-linked with one cytosine residue of domain V RNA through its Asn152 residue. (A) MS spectrum of the signal at m/z 1317.78, derived after addition with a cytosine (C) residue (molecular weight, 305) from the “parent ion” at m/z 1013.48 (VGDANPALQK) (M+H)+. (B) The sequence of the corresponding peptide, with a scheme of the y and b fragmentation series. The asparagine residue within the box is cross-linked with the C moiety. (C) MS/MS spectrum of modified VGDANPALQK (m/z = 1317.78, i.e., attached with C) recorded with a 2,5-dihydroxy benzoic acid matrix. The spectra show an almost complete series of y-type and b-type fragments (b5/y5 to b9/y9 shown). Ion b5 is attached with a C residue, as revealed by the m/z value, while y6 is also attached with a C residue, so Asn152 is cross-linked with the C residue of domain V. (D) Complete list of b and y ions is presented here. b1 to b4 and y1 to y5 indicate unmodified ions. b5* to b9* and y6* to y9* represent modified ions attached with a C residue (the asterisk is used to depict modified ions).
Identification of specific amino acids in BCA and lysozyme that interact with nucleotides in domain V RNA.
Amino acids of BCA which interact with domain V RNA were identified through MS/MS studies of the peptides. Interacting amino acids are shown in boldface in the 1 SHHWGYGK 8, 58 MVNNGHSFNVEYDDSQDK 75, 80 DGPLTGTYR 88, 148 VGDANPALQK 157, and 253 QVRGFPK 259 stretches. The details of MS/MS results are shown for 148 VGDANPALQK 157 only (Fig. 3). In the MS/MS spectra, the modified (i.e., attached with cytosine residue) b1 to b4 ions are absent but modified forms of b5, b6, b7, and b9 are present (Fig. 3C). A scan of y ions revealed that only y6, y8, and y9 were modified (Fig. 3C). The figure clearly shows that asparagine 152 is the amino acid in this oligopeptide that comes in contact with domain V RNA during folding. For lysozyme, the interacting amino acids are shown in boldface in the 6 CELAAAMK 13, 14 RHGLDNYR 21, 34 FESNFNTQATNR 45, 62 WWCNDGR 68, and 115 CKGTDVQAWIR 125 stretches. Detailed information on the amino acid residues and the nucleotides with which they interact is shown in Table 1.
TABLE 1.
Contact sites between unfolded BCA or lysozyme and domain V RNA during foldinga
| Protein | Peptide sequence | Nucleotide sequence | Cross-linked nucleotide in domain V RNA |
|---|---|---|---|
| BCA | 1 SHHWGYGK 8 | 2477 UACUUG 2472 | U-2473 |
| 58 MVNNGHSFNVEYDDSQDK 75 | 2495 GGUUUG 2490 | U-2492/2491 | |
| 80 DGPLTGTYR 88 | 2589 AGAUUU 2584 | A-2587 | |
| 148 VGDANPALQK 157 | 2555 UUGUCG 2550 | C-2551 | |
| 253 QVRGFPK 259 | 2564 AUUUAC 2559 | A-2560 | |
| Lysozyme | 6 CELAAAMK 13 | 2589 AGAUUU 2584 | A-2587 |
| 14 RHGLDNYR 21 | 2495 GGUUUG 2490 | U-2492/U-2491 | |
| 34 FESNFNTQATNR 45 | 2477 UACUUG 2472 | U-2473 | |
| 62 WWCNDGR 68 | 2555 UUGUCG 2550 | C-2551 | |
| 115 CKGTDVQAWIR 125 | 2564 AUUUAC 2559 | A-2560 |
Interacting amino acids and nucleotides are represented as bold one-letter code.
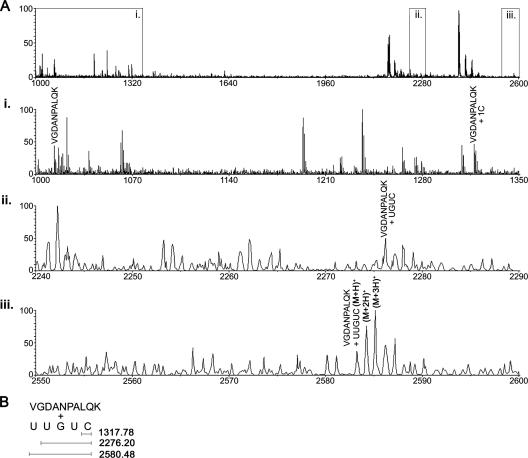
The specific combinations of nucleotides and amino acids involved in the interaction were figured out by combining the results from primer extension, MS, and MS/MS experiments. Figure 4A shows spectral ranges containing the peptide 148 VGDANPALQK 157 at m/z 1012.48 and its attachment with the cytosine residue at m/z 1317.78 (Fig. 4A, i). As we went forward to check whether the same peptide was attached with larger RNA segments, we found it to be cross-linked with two more stretches of RNA, UGUC (Fig. 4A, ii) and UUGUC (Fig. 4A, iii). From all of these observations, the deduced oligonucleotide sequence of domain V RNA, which is significant for binding of this peptide, is shown in Fig. 4B. The cytosine residue present at the 5′ end of the 3′ UUGUC 5′ stretch is responsible for binding with the peptide VGDANPALQK. A complete analysis of interaction of domain V RNA with all other peptide stretches of BCA and lysozyme is given in Table 1. Comparing the amino acid-nucleotide interactions, we see that two of these combinations are the same for the two proteins.
FIG. 4.
MS analysis of VGDANPALQK cross-link to show the RNA sequence where it binds through the cytosine residue. (A) Spectral range (m/z 1000 to 2600) which includes the peptide stretch alone (m/z 1012.48) and its different cross-linked form. (i) The VGDANPALQK peptide only and the same peptide cross-linked with one C residue are shown. (ii) MS spectrum of the signal at m/z 2276.20, derived from the VGDANPALQK peptide, where it remains attached with the UGUC nucleotide stretch of domain V. (iii) The same peptide, while attached to the UUGUC nucleotide stretch of domain V, gives a signal at m/z 2580.48. (B) The deduced oligonucleotide sequence of domain V RNA is shown; the last C residue of the 3′ UUGUC 5′ stretch interacts with the peptide.
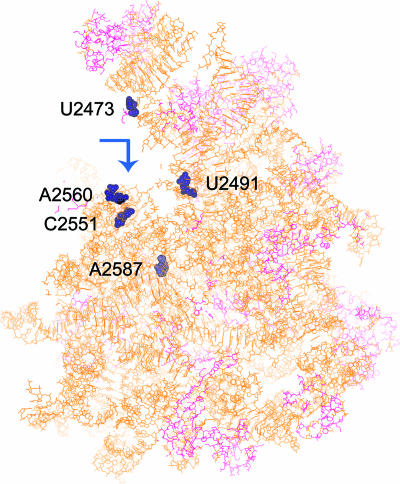
The five nucleotides mentioned in the table and now shown in the crystal structure of the E. coli 50S ribosomal subunit (PDB identifier 2AW4) in Fig. 5 belong to the sets shown as reverse transcriptase stops in the primer extension experiment (Fig. 2). As has been seen by others, the stops occur at a few nucleotides just downstream of the actual nucleotide with which the protein binds (39). Interestingly, most of the interacting amino acids of both the proteins are in the loop regions (Fig. 6), although the sequences in the two cases are not identical. Moreover, the identified peptide stretches mapped on the surfaces of the crystal structures of BCA (PDB identifier 1V9E) and lysozyme (PDB identifier 1GXV). Thus, specific regions of unfolded proteins seem to be involved in interaction with domain V for attaining the active (folded) form.
FIG. 5.
Atomic resolution crystal (3.5 Å) structure of the E. coli 50S ribosomal subunit (PDB identifier 2AW4) showing the nucleotides involved in protein folding. Nucleotides U2473, U2491, C2551, A2560, and A2587, which interact with the unfolded protein, are situated at the entry mouth (shown by an arrow) of the tunnel and are depicted as CPK models. The figure was created using the BIOSYM package (Insight II; Biosym/MSI).
FIG. 6.
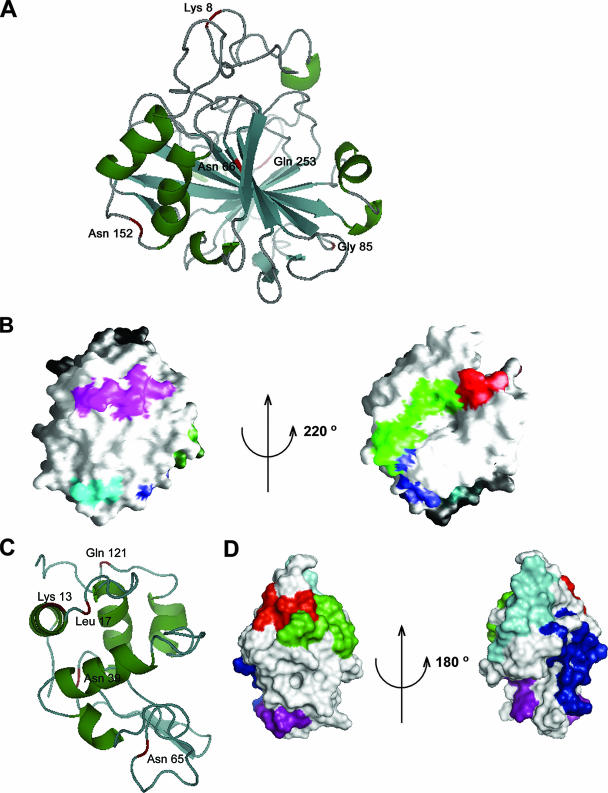
Surface residues of BCA are shown which interact with domain V RNA. (A) Ribbon diagram of BCA (PDB identifier 1V9E) showing five residues (labeled and marked in red) that interact with RNA during folding. Four of these amino acids are located in the loop regions. The cartoon was drawn using the PyMOL software program. (B) Surface view (using the GRASP software program) of two approximately opposite faces of the BCA molecule where the interacting peptide stretches 1 SHHWGYGK 8 (red), 58 MVNNGHSFNVEYDDSQDK 75 (green), 80 DGPLTGTYR 88 (blue), 148 VGDANPALQK 157 (purple), and 253 QVRGFPK 259 (cyan) are located. (C) Ribbon diagram (using PyMOL) of lysozyme (PDB identifier 1GXV) showing five residues (labeled and marked in red) that interact with RNA during folding. (D) Surface view (using PyMOL) of two opposite faces of lysozyme where the interacting peptide stretches 6 CELAAAMK 13 (red), 14 RHGLDNYR 21 (green), 34 FESNFNT QATNR 45 (blue), 62 WWCNDGR 68 (purple), and 115 CKGTDVQAWIR 125 (cyan) are marked.
DISCUSSION
The protein folding activity of domain V RNA has added a new perspective to the protein folding problem. Our present finding of the involvement of a unique set of nucleotides in PTC with amino acids of different proteins is a striking observation. The amino acids involved, although not identical in different proteins, are found to be the fairly conserved ones. Our results point toward a chemistry built up in the course of evolution that needs to be explained.
The structural overlap of the two functions, protein synthesis and folding, on the PTC (8, 25-27, 30) suggests that they coevolved in the RNA world. The secondary structure of domain V has been preserved throughout evolution, and some of the bases in its large central loop are invariant (16). We find that most of the bases that contact proteins are in the loops and bulges, which agrees with the current view on RNA-protein interaction.
The specific nucleotides in PTC implicated in protein folding map on the crystal structure of the 50S subunit in the cavity at the entry mouth of the exit tunnel that goes through the 50S subunit (Fig. 5). We measured the affinity constants of unfolded BCA for 70S and 50S particles to be 9 × 107 M−1 and 9 × 108 M−1, respectively. The corresponding values for native BCA were 2 × 106 M−1 and 4 × 106 M−1, respectively (unpublished data). Therefore, the newly synthesized proteins, released from the 70S ribosome, have a strong preference to bind to the 50S subunit. In fact, in recent publications (4, 5), we have shown that full-length proteins remain associated with free 50S subunits when the 70S ribosome was used for folding in vitro (5) and when the proteins were synthesized in a cell-free E. coli translation mix or in live E. coli cells (4). These results suggest that PTC-mediated protein folding could be physiologically relevant.
The binding of full-length nascent proteins to the 50S subunit must follow the splitting of the 70S ribosome, which takes place after complete synthesis of the protein. The proteins most likely exit in a largely unfolded state from the exit tunnel because it is too narrow to accommodate proteins with tertiary folds (2, 26). Upon release from the tunnel, the proteins can bind to free 50S subunits in the cytosol. Alternatively, the nascent protein could bind to the PTC of the ribosome from which it was synthesized, but this would require going around the 50S subunit to the PTC at the tunnel entry point, which seems unlikely.
Another possibility, that all proteins might not exit from the tunnel, is suggested from the following observations. The folding of globin (heme binding site formation) takes place on the PTC, and this intermediate is too large to go through the tunnel and has to leave via the interface between the two subunits (22). Some nascent peptide chains cross-link to 16S rRNA (10). The tail spike protein of phage P22 remains associated with the 30S subunit after the subunits dissociate (12). The nascent proteins could also get distributed in branches from the exit tunnel seen in the rear side of the large subunit (36). In a cryoelectron microscopy study, three nascent proteins, Ig1, Ig2, and green fluorescent protein, were found to be largely concentrated in the entry cavity of the tunnel (PTC region) in E. coli ribosomes stalled following translation (18). In these and in many earlier studies (6), the full-length protein is found to exit from a point which is at a distance from the PTC. Whether the exit is from within or outside the tunnel thus remains unsettled.
Earlier we showed that folding of a number of proteins, including β-galactosidase and the DnaK chaperone in vitro and in vivo and firefly luciferase translated in vitro, could be suppressed by antibiotics that bind to domain V (4, 9, 17). These results lead us to believe that the folding of full-length proteins by domain V RNA in vitro is a good mimicry of the folding in vivo.
In the primer extension experiments, we found that in most cases the nucleotide bases in PTC interacted directly with the amino acids of unfolded proteins; only in a few cases could the interaction be through the phosphate moiety of the nucleotides (see Table S2 in the supplemental material). Some of the ambiguities in the results of primer extension experiments, like the binding of folding proteins to more than one (consecutive) nucleotide, were cleared by the MS studies. Among the amino acids of the two proteins (BCA and lysozyme) studied here, asparagine interacted with RNA the most frequently, with the other amino acids being lysine, glutamine, leucine, and glycine. This agrees with the list of amino acids that are prevalent among those interacting with nucleotides of RNA (21, 32). We have not detected any negatively charged amino acid interacting with RNA.
The amino acids that interact with domain V were found to be present mostly on random coils on the surfaces of the crystal structures of globular proteins (Fig. 6). The forces and interactions working on the polypeptide when it folds in an aqueous environment can give it the right shape if topological and steric problems are overcome. We can imagine three major steps where domain V RNA could help.
(i) A major deficit that must be overcome during folding is the large positive-configurational entropy of random coils (15). Reducing this entropy by clamping the amino acids in random coils on the outside of the protein could be the major contribution of domain V.
(ii) Another major problem is the unfavorable standard free energy of transfer associated with dehydration of hydrophobic functional groups as they are pulled into the interior of folded polypeptides (33). This effect can be largely modulated by slowing down the process by reducing the temperature to give the surrounding water a more organized structure. We found that folding by ribosome and domain V RNA is very efficient at low temperatures. It is general knowledge that the proteins retain their native conformation much longer at low temperatures.
(iii) A third advantage is trapping of the folding protein at the entry mouth of the 50S ribosomal tunnel (18) through the external random coils with the specific nucleotides of PTC. This would considerably reduce the overall entropy of the system.
Once these disruptive processes are tamed, the internalized secondary structures might engage in a jigsaw puzzle to proceed toward native conformation. We are trying to delete/mutate the amino acids which bind to specific nucleotides to get more insight into the process of ribosome-assisted protein folding. From studies of only two proteins, we found two identical amino acid-nucleotide pairs out of five such pairs. Whether this is fortuitous or not would be made evident by studying such interactions with a few more proteins.
Supplementary Material
Acknowledgments
We thank Dhruba Chattoraj (NIH) for many thoughtful comments.
This research was supported by grants from DBT, CSIR, FIST, UGC-DSA, and UGC-COSIST. D.S. and A.D. are CSIR research fellows.
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Argent, R. H., A. M. Parrott, P. J. Day, L. M. Roberts, P. G. Stockley, J. M. Lord, and S. E. Radford. 2000. Ribosome-mediated folding of partially unfolded ricin A-chain. J. Biol. Chem. 2759263-9269. [DOI] [PubMed] [Google Scholar]
- 2.Ban, N., P. Nissen, J. Hansen, M. Capel, P. B. Moore, and T. A. Steitz. 1999. Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit. Nature 400841-847. [DOI] [PubMed] [Google Scholar]
- 3.Barsnes, H., S. O. Mikalsen, and I. Eidhammer. 2006. MassSorter: a tool for administrating and analyzing data from mass spectrometry experiments on proteins with known amino acid sequences. BMC Bioinform. 742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu, A., D. Samanta, A. Bhattacharya, A. Das, D. Das, and C. Dasgupta. 2008. Protein folding following synthesis in vitro and in vivo: association of newly synthesized protein with 50S subunit of E. coli ribosome. Biochem. Biophys. Res. Commun. 366592-597. [DOI] [PubMed] [Google Scholar]
- 5.Basu, A., D. Samanta, D. Das, S. Chowdhury, A. Bhattacharya, J. Ghosh, A. Das, and C. Dasgupta. 2008. In vitro protein folding by E. coli ribosome: unfolded protein splitting 70S to interact with 50S subunit. Biochem. Biophys. Res. Commun. 366598-603. [DOI] [PubMed] [Google Scholar]
- 6.Bernabeu, C., and J. A. Lake. 1982. Nascent polypeptide chains emerge from the exit domain of the large ribosomal subunit: immune mapping of the nascent chain. Proc. Natl. Acad. Sci. USA 793111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattopadhyay, S., B. Das, A. K. Bera, D. Dasgupta, and C. Dasgupta. 1994. Refolding of denatured lactate dehydrogenase by Escherichia coli ribosomes. Biochem. J. 300717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyay, S., B. Das, and C. Dasgupta. 1996. Reactivation of denatured proteins by 23S ribosomal RNA: role of domain V. Proc. Natl. Acad. Sci. USA 938284-8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chattopadhyay, S., S. Pal, D. Pal, D. Sarkar, S. Chandra, and C. Das Gupta. 1999. Protein folding in Escherichia coli: role of 23S ribosomal RNA. Biochim. Biophys. Acta 1429293-298. [DOI] [PubMed] [Google Scholar]
- 10.Choi, K. M., J. F. Atkins, R. F. Gesteland, and R. Brimacombe. 1998. Flexibility of the nascent polypeptide chain within the ribosome—contacts from the peptide N-terminus to a specific region of the 30S subunit. Eur. J. Biochem. 255409-413. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury, S., S. Pal, J. Ghosh, and C. DasGupta. 2002. Mutations in domain V of the 23S ribosomal RNA of Bacillus subtilis that inactivate its protein folding property in vitro. Nucleic Acids Res. 301278-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark, P. L., and J. King. 2001. A newly synthesized, ribosome-bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates. J. Biol. Chem. 27625411-25420. [DOI] [PubMed] [Google Scholar]
- 13.Das, B., S. Chattopadhyay, A. K. Bera, and C. Dasgupta. 1996. In vitro protein folding by ribosomes from Escherichia coli, wheat germ and rat liver: the role of the 50S particle and its 23S rRNA. Eur. J. Biochem. 235613-621. [DOI] [PubMed] [Google Scholar]
- 14.Das, B., S. Chattopadhyay, and C. Das Gupta. 1992. Reactivation of denatured fungal glucose 6-phosphate dehydrogenase and E. coli alkaline phosphatase with E. coli ribosome. Biochem. Biophys. Res. Commun. 183774-780. [DOI] [PubMed] [Google Scholar]
- 15.Dill, K. A. 1985. Theory for the folding and stability of globular proteins. Biochemistry 241501-1509. [DOI] [PubMed] [Google Scholar]
- 16.Douthwaite, S. 1992. Functional interactions within 23S rRNA involving the peptidyl transferase center. J. Bacteriol. 1741333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, J., A. Basu, S. Pal, S. Chowdhuri, A. Bhattacharya, D. Pal, D. K. Chattoraj, and C. DasGupta. 2003. Ribosome-DnaK interactions in relation to protein folding. Mol. Microbiol. 481679-1692. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, R. J., P. Fucini, S. Connell, S. D. Fuller, K. H. Nierhaus, C. V. Robinson, C. M. Dobson, and D. I. Stuart. 2004. Three-dimensional structures of translating ribosomes by Cryo-EM. Mol. Cell 1457-66. [DOI] [PubMed] [Google Scholar]
- 19.Gregory, S. T., and A. E. Dahlberg. 1999. Mutations in the conserved P loop perturb the conformation of two structural elements in the peptidyl transferase center of 23 S ribosomal RNA. J. Mol. Biol. 2851475-1483. [DOI] [PubMed] [Google Scholar]
- 20.Hardesty, B., and G. Kramer. 2001. Folding of a nascent peptide on the ribosome. Prog. Nucleic Acid Res. Mol. Biol. 6641-66. [DOI] [PubMed] [Google Scholar]
- 21.Jones, S., D. T. Daley, N. M. Luscombe, H. M. Berman, and J. M. Thornton. 2001. Protein-RNA interactions: a structural analysis. Nucleic Acids Res. 29943-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komar, A. A., A. Kommer, I. A. Krasheninnikov, and A. S. Spirin. 1997. Cotranslational folding of globin. J. Biol. Chem. 27210646-10651. [DOI] [PubMed] [Google Scholar]
- 23.Kudlicki, W., A. Coffman, G. Kramer, and B. Hardesty. 1997. Ribosomes and ribosomal RNA as chaperones for folding of proteins. Fold. Des. 2101-108. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn-Holsken, E., C. Lenz, B. Sander, R. Luhrmann, and H. Urlaub. 2005. Complete MALDI-ToF MS analysis of cross-linked peptide-RNA oligonucleotides derived from nonlabeled UV-irradiated ribonucleoprotein particles. RNA 111915-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moazed, D., and H. F. Noller. 1991. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc. Natl. Acad. Sci. USA 883725-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289920-930. [DOI] [PubMed] [Google Scholar]
- 27.Noller, H. F., V. Hoffarth, and L. Zimniak. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 2561416-1419. [DOI] [PubMed] [Google Scholar]
- 28.O'Connor, M., and A. E. Dahlberg. 1993. Mutations at U2555, a tRNA-protected base in 23S rRNA, affect translational fidelity. Proc. Natl. Acad. Sci. USA 909214-9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Connor, M., and A. E. Dahlberg. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 254838-847. [DOI] [PubMed] [Google Scholar]
- 30.Pal, D., S. Chattopadhyay, S. Chandra, D. Sarkar, A. Chakraborty, and C. Das Gupta. 1997. Reactivation of denatured proteins by domain V of bacterial 23S rRNA. Nucleic Acids Res. 255047-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal, S., S. Chandra, S. Chowdhury, D. Sarkar, A. N. Ghosh, and C. D. Gupta. 1999. Complementary role of two fragments of domain V of 23 S ribosomal RNA in protein folding. J. Biol. Chem. 27432771-32777. [DOI] [PubMed] [Google Scholar]
- 32.Phipps, K. R., and H. Li. 2007. Protein-RNA contacts at crystal packing surfaces. Proteins 67121-127. [DOI] [PubMed] [Google Scholar]
- 33.Privalov, P. L., and G. I. Makhatadze. 1993. Contribution of hydration to protein folding thermodynamics. II. The entropy and Gibbs energy of hydration. J. Mol. Biol. 232660-679. [DOI] [PubMed] [Google Scholar]
- 34.Rhode, B. M., K. Hartmuth, H. Urlaub, and R. Luhrmann. 2003. Analysis of site-specific protein-RNA cross-links in isolated RNP complexes, combining affinity selection and mass spectrometry. RNA 91542-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanyal, S. C., S. Pal, S. Chowdhury, and C. DasGupta. 2002. 23S rRNA assisted folding of cytoplasmic malate dehydrogenase is distinctly different from its self-folding. Nucleic Acids Res. 302390-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenson, T., and M. Ehrenberg. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108591-594. [DOI] [PubMed] [Google Scholar]
- 37.Thiede, B., H. Urlaub, H. Neubauer, G. Grelle, and B. Wittmann-Liebold. 1998. Precise determination of RNA-protein contact sites in the 50S ribosomal subunit of Escherichia coli. Biochem. J. 33439-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urlaub, H., K. Hartmuth, and R. Luhrmann. 2002. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods 26170-181. [DOI] [PubMed] [Google Scholar]
- 39.Urlaub, H., K. Hartmuth, S. Kostka, G. Grelle, and R. Luhrmann. 2000. A general approach for identification of RNA-protein cross-linking sites within native human spliceosomal small nuclear ribonucleoproteins (snRNPs). Analysis of RNA-protein contacts in native U1 and U4/U6.U5 snRNPs. J. Biol. Chem. 27541458-41468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.