Abstract
XylS, an AraC family protein, activates transcription from the benzoate degradation pathway Pm promoter in the presence of a substrate effector such as 3-methylbenzoate (3MB). We developed a procedure to obtain XylS-enriched preparations which proved suitable to analyze its activation mechanism. XylS showed specific 3MB-independent binding to its target operator, which became strictly 3MB dependent in a dimerization-defective mutant. We demonstrated that the N-terminal domain of the protein can make linker-independent interactions with the C-terminal domain and inhibit its capacity to bind DNA. Interactions are hampered in the presence of 3MB effector. We propose two independent roles for 3MB in XylS activation: in addition to its known influence favoring protein dimerization, the effector is able to modify XylS conformation to trigger N-terminal domain intramolecular derepression. We also show that activation by XylS involves RNA polymerase recruitment to the Pm promoter as demonstrated by chromatin immunoprecipitation assays. RNA polymerase switching in Pm transcription was reproduced in in vitro transcription assays. All σ32-, σ38-, and σ70-dependent RNA polymerases were able to carry out Pm transcription in a rigorous XylS-dependent manner, as demonstrated by the formation of open complexes only in the presence of the regulator.
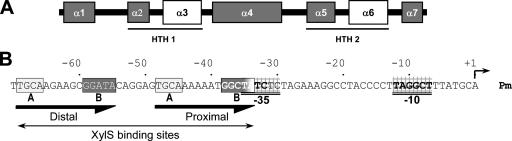
XylS belongs to the AraC family of transcription regulators (14, 57). This family includes a large number of activators that control expression of genes involved in virulence, stress, and metabolism. The DNA-binding domain of the family members is highly conserved and spans a 100-amino-acid stretch, generally at the C-terminal end of the protein (14, 74). Most family members also contain an effector-binding domain which modulates their activity and carries dimerization determinants (6, 48, 52, 53, 57). Early work based on structure prediction analysis allowed the definition of two helix-turn-helix (HTH) DNA binding motifs within the conserved DNA-binding domain (14). Direct structural analysis of most proteins of the family has been hampered by protein insolubility. However, MarA and Rob proteins were crystallized in complex with their cognate binding sites in the DNA. The X-ray crystallographic data confirmed that the conserved DNA-binding domain is composed of seven α-helices folding in two HTH motifs (Fig. 1A). The two HTH recognition helices, α3 and α6, bind two adjacent segments of the major groove (34, 59). Interestingly, both proteins crystallized as monomers.
FIG. 1.
(A) Schematic representation of XylS-C structure. The predicted α-helixes in the sequence are depicted as gray boxes. The relative sizes of the different helixes are drawn to scale. DNA-contact helixes α3 and α6 are in white. (B) Pm promoter sequence organization. The bold arrows indicate the two XylS binding sites (proximal and distal), each composed of conserved A1/A2 and B1/B2 boxes. The −10 and −35 hexamers are in bold and double-underlined. A right-angled arrow indicates the transcription initiation site.
XylS is the transcriptional activator of Pm, the promoter that controls expression of the meta-cleavage pathway for benzoate and alkylbenzoate catabolism coded by the Pseudomonas putida TOL plasmid (40, 56). Two factors modulate XylS-dependent transcription from Pm: the presence of effector molecules such as the pathway substrate 3-methylbenzoate (3MB), which triggers XylS activation, and the intracellular levels of XylS regulator. In fact, in most conditions, XylS is synthesized at low levels and is activated by the effector to promote transcription from Pm. However, when XylS is overproduced from the inducible PS1 promoter in the presence of toluene, it activates Pm in the absence of 3MB (12, 17, 56).
As with other members of the family, the DNA-binding domain of XylS is located at the C-terminal end of the protein, connected by a linker to the 200-amino-acid N-terminal end. Genetic evidence suggests that this domain is involved in effector recognition (47, 62). In AraC, effector binding has major consequences on the protein structure and function. This regulator operates as a dimer in which a flexible arm formed by the first 20 amino acids of the N-terminal domain are essential for the protein switch between two conformations with opposite functions (37, 68) (see below). The molecular consequences of effector binding in XylS are unknown, and the oligomeric state of the XylS protein has not been determined directly. This protein, like many AraC family members, is insoluble at higher concentrations, and accurate biochemical determinations have proven unattainable. However, indirect evidence suggests that the XylS N-terminal domain is able to dimerize in a process that, at low protein concentrations, depends on the presence of the effector (61).
XylS recognizes two 15-bp direct repeats (TGCA-N6-GGNTA), each consisting of two half-sites: a 5′ box A1/A2 (TGCA) and a 3′ box B1/B2 (GGNTA) (Fig. 1B) (13, 16, 31). The arrangement of the two repeats is such that the proximal XylS binding site overlaps the RNA polymerase −35 binding box by 2 bp (15) (Fig. 1B). According to Busby and Ebright, the Pm promoter can be considered a class II promoter (5). At these promoters, regulators activate transcription by establishing multiple interactions with the RNA polymerase α and σ subunits. Contacts with the C-terminal domain of the RNA polymerase α subunit have been implicated in transcription activation by a number of AraC family members (27-29, 36, 63, 66). Some AraC family proteins also require specific amino acid interactions with RNA polymerase σ70 region 4 to activate transcription (3, 20, 35). Interestingly, transcription from Pm promoter uses the RNA polymerase holoenzyme containing σ32 (Eσ32) in exponential phase but Eσ38 in stationary phase. Thus, XylS must be able to contact RNA polymerase holoenzyme with different sigma factors.
Although the activation mechanisms used by AraC family members are poorly understood, some points have been clarified. MarA and SoxS proteins solely contain the DNA-binding domain and lack the effector response domain. They are synthesized de novo in response to their inducing signals, resulting in a 10-fold increase in intracellular levels, which allows activation at their target promoters (22). Upon removal of the inducing signal, activator levels decrease through protein degradation and return to basal levels (22, 28). In contrast, some effector-responsive AraC family members containing two functional domains influence DNA topology: in the absence of effectors, AraC and MelR bind two distant sites in the operator, giving rise to DNA loops that repress gene expression (30, 42). Effector binding leads to a conformational change involving occupancy of adjacent DNA binding sites; the DNA loops are thus dismantled, and gene expression is induced from the corresponding promoters (30, 37, 42, 77). In AraC and MelR the RNA polymerase is recruited to the target promoters in the presence of the corresponding effector (arabinose or melibiose, respectively), stimulating isomerization to open complex (19, 20, 81).
Despite our substantial knowledge of XylS and other members of the AraC family of transcriptional activators, the molecular events underlying XylS activation remain unsolved. The activity of this regulator shares features of both types of family proteins. As MelR and AraC, it is activated by the presence of an effector, but it is also able to activate transcription when overproduced through a physiological regulatory cascade (55). In this work we investigated the processes of XylS binding to DNA and protein dimerization, and we analyzed the role of the effector 3MB. Our aim was to establish connections between these processes to shed light on the mechanism of XylS activation. To this end, we used extract preparations enriched in XylS, and we were able to describe the different molecular events leading to promoter activation, where the effector plays a dual role: 3MB, which was previously shown to favor XylS dimerization (61), is also required to release intramolecular inhibition.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA techniques.
The bacterial strains and plasmids used in this work are listed in Table 1. Bacterial strains were routinely grown at 30°C and 200 rpm in Luria-Bertani (LB) medium as described before (9). Growth was determined turbidometrically at 660 nm. The plasmids pCMX2, pLOW2, pJLR100, pJLR107, pERD103, and pMD::Pm245 have been described previously (Table 1). All DNA manipulations were done according to standard procedures (1) or to the manufacturer's recommendations.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| CC118 | recA1 supE44 endA1 hsdR17 (rK− mK−) gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+lacIqlacZΔM15] | 80 |
| CC118λPm::lacZ | CC118, lysogenized with a λPm::lacZ fusion | 33 |
| BL21(DE3) | Carries T7 RNA polymerase under the control of lacUV5 promoter | Novagen |
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U196 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 25 |
| Plasmids | ||
| pJLR100 | Apr, Pm cloned in pEMBL9 | 58 |
| pCMX2 | Tcr, pSELECT derivative containing the wild-type xylS gene | 38 |
| pERD103 | Kmr, derivative containing the wild-type xylS gene | 56 |
| pLOW2 | Kmr, pACYC177 derivative, low-copy-no. cloning vector | 26 |
| pLOW2::XylS | pLOW2 derivative containing the xylS gene cloned as an EcoRI-XbaI fragment | 62 |
| pJLR107 | Apr, Pm::lacZ in pMD1405 | 58 |
| pMD::Pm245 | Apr, Pm245::lacZ in pMD1405 | 16 |
| pET16b::XylS-C | Apr, pET16b derivative used to produce His-tagged XylS C-terminal domain | Domínguez-Cuevas et al., unpublished |
| pET16b::rpoS | Apr, pET16b derivative used to produce His-tagged P. putida σ38 | This work |
| pET16b::rpoH | Apr, pET16b derivative used to produce His-tagged P. putida σ32 | This work |
| pET16b::XylS-N | Apr, pET16b derivative used to produce His-tagged XylS N-terminal domain | This work |
| pGP1-2 | Kmr, contains an inducible T7 RNA polymerase gene | 70 |
| pBBR1::XylS-C | Gmr, pBBR1MSC5 derivative containing XylS-C | This work |
Apr, Kmr, and Tcr indicate resistance to ampicillin, kanamycin, and tetracycline, respectively.
Preparation of cleared-out extracts enriched in XylS.
Escherichia coli CC118 bearing pCMX2 or pLOW2::xylS was grown overnight in LB medium with tetracycline (10 μg/ml) or kanamycin (25 μg/ml) at 30°C and 200 rpm. Flasks containing 500 ml of fresh medium were inoculated with an aliquot fraction of these precultures and were incubated at 30°C. When the optical density at 600 nm reached 0.6, cultures were supplemented with 1 mM 3MB or left untreated, and incubation continued for 3 h. The cultures were then harvested by centrifugation, and cell pellets were stored at −70°C. Crude extracts were prepared by resuspending the pellet in 3 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 4 mM β-mercaptoethanol, 2 mM EDTA, 1× Complete protease inhibitor mixture [Roche Applied Science]), followed by cell disruption by sonication. The clear supernatant was filtered through a 0.45-μm-pore-size nylon membrane and loaded onto a 1-ml heparin column (HiTrap Heparin HP; Amersham Biosciences) preequilibrated with buffer A (10 mM sodium phosphate, pH 7, 4 mM β-mercaptoethanol). When extracts were prepared from 3MB-grown cells, this step was carried out in the presence of 3MB, and the resulting protein solution was used for electrophoretic mobility shift assay (EMSA) in the presence of 1 mM 3MB. When extracts were prepared from cells grown in the absence of 1 mM 3MB, this effector was omitted in subsequent steps. Unbound material was collected, and the column was washed with buffer A until nonspecifically bound material was removed. Specifically bound proteins were eluted from the column with buffer A supplemented with 1 M NaCl. The protein concentration in the fractions was determined according to Bradford (4).
Western blots.
Cell extract preparations obtained as above or purified XylS protein were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) (12.5%) and transferred to nitrocellulose membranes. Membranes were blocked for 3 h at room temperature with 5% nonfat dry milk in phosphate-buffered saline. Blots were incubated at 4°C overnight with a 1/1,000 dilution of polyclonal antiserum against XylS (76). Blots were washed with phosphate-buffered saline solution and incubated with goat anti-rabbit immunoglobulin G (H+L) conjugated with horseradish peroxidase (1:1,000 dilution) for 1 h (Caltag Laboratories). The blots were developed with the SuperSignal West Dura Extended Duration Substrate (Pierce). Chemiluminescent blots were exposed in a Chemi-Doc (Bio-Rad) luminometer.
Overexpression and purification of His-tagged XylS C-terminal and N-terminal domains.
The 400-bp DNA fragment covering the XylS C-terminal domain (XylS residues 196 to 321) was amplified by PCR from the pERD103 plasmid using primers XylSCNdeI (5′-GGAATTCCATATGCTGGGCAGCAATGTCAGC-3′) and XylSXhoI (5′-CCGCTCGAGTCAAGCCACTTCCTTTTTGC-3′). The PCR product was digested with NdeI and XhoI enzymes and subsequently cloned into the pET16b vector (Novagen). The histidine-tagged XylS-C terminal domain (hereafter, Xyls-C) was overexpressed and purified as described previously (P. Domínguez-Cuevas, J. L. Ramos, and S. Marqués, unpublished data). Electrophoresis of 10 μg of this protein preparation gave a single Coomassie-stained band. The 643-bp DNA fragment covering XylS-N terminal domain (XylS residues 1 to 207) was amplified by PCR from plasmid pERD103 using primers XylSNdeI (5′-GGAATTCCATATGGATTTTTGCTTATTGAACGAG-3′) and XylSNterXhoI (5′-CCGCTCGAGTCAGCTGAAAATTTCACGGCTGAC-3′). The PCR product was digested with NdeI and XhoI enzymes and subsequently cloned into the pET16b vector (Novagen). Freshly transformed BL21(DE3) cells harboring the pET16b::XylS-N plasmid were grown in 500 ml of 2× YT (yeast extract and tryptone) medium (64) at 30°C until turbidity at 660 nm reached 0.5. At this point the culture was transferred to 16°C, and the addition of 0.1 mM isopropyl-beta-d-thiogalactopyranoside (IPTG) allowed the induction of fusion proteins during the subsequent 3 to 4 h of incubation, after which cells were pelleted and frozen. The cells were resuspended in 50 ml of lysis buffer (30 mM Tris-HCl, pH 8.8, 300 mM NaCl, 0.1 mM EDTA, 2.5 mM 2-mercaptoethanol, 10% [vol/vol] glycerol, 10 mM imidazole, 0.5% Triton X-100, and 1 mM Complete protease inhibitor mixture [Roche Applied Science]) and disrupted in a French pressure cell. After centrifugation at 22,500 × g for 45 min, the inclusion body was resuspended in 60 ml of solubilization buffer (30 mM Tris-HCl, pH 8.8, 500 mM NaCl, 0.1 mM EDTA, pH 8, 2.5 mM 2-mercaptoethanol, 10% [vol/vol] glycerol, 10 mM imidazole, 0.05% Triton X-100, and 6 M guanidine-HCl). After centrifugation at 30,000 × g for 60 min, the supernatant fraction was passed through a 0.45-μm-pore-size filter and loaded onto a 5-ml Ni-agarose column (Amersham Biosciences) preequilibrated with solubilization buffer. Nonspecifically bound material was washed with five volumes of preequilibration solubilization buffer, and the His-tagged XylS N-terminal domain (hereafter, XylS-N) was eluted from the column at about 400 mM imidazole in a 30-ml elution buffer gradient from 0 to 1 M imidazole. The purified XylS-N was refolded by sequential dialysis against dialysis buffers (30 mM Tris-HCl, pH 8.8, 300 mM KCl, 0.1 mM EDTA, 2 mM dithiothreitol [DTT], 10% [vol/vol] glycerol, 1 mM glutathione-0.2 mM glutathione disulfide) and decreasing urea concentrations from 4 M to 0 M. Electrophoresis of 10 μg of the resulting protein solution gave a single Coomassie-stained band.
EMSAs.
The 100-bp DNA fragments containing the wild-type or mutant xylS binding site (positions −110 to −10 of the Pm promoter) were amplified by PCR with primers Pm3 (5′-CTGCAGTGTCCGGTTTGATAGGG-3′) and Pm4 (5′-CCTAAGGGGTAGGCCTTTCTAG-3′). The PCR products were isolated from agarose gels and end labeled with [γ-32P]ATP as described previously (72). The indicated amount of XylS-enriched extracts and a 1 nM concentration of end-labeled DNA fragments were mixed and incubated at 30°C for 15 min in 10 μl of binding buffer (Tris-glycine buffer [25 mM Tris-HCl, 200 mM glycine, pH 8.6], 200 mM NaCl, 4 mM β-mercaptoethanol, 4 mM MgCl2, 4 mM EDTA), supplemented when indicated with 1 mM 3MB. Samples were loaded onto a 4.5% nondenaturing polyacrylamide gel and electrophoresed at 50 V in Tris-glycine buffer (25 mM Tris-HCl, 200 mM glycine, pH 8.6) for 2 h at 4°C. The gels were dried and visualized by exposure to phosphorimager screens. The results were analyzed with Molecular Imager FX equipment and the QuantityOne software (Bio-Rad, Madrid, Spain).
Overexpression and purification of Pseudomonas putida sigma factors.
The 874-bp and 1,029-bp DNA fragments containing the rpoH and rpoS genes, respectively, were amplified by PCR from P. putida KT2440 chromosomal DNA using primers rpoHNdeI (5′-GGAATTCCATATGACCACATCGTTGCAACC-3′), rpoHXhoI (5′-CCGCTCGAGTCAGGCAGCGATCAGTGCC-3′), rpoSNdeI (5′-GGAATTCCATATGGCTCTCAGTAAAGAAGT-3′), and rpoSXhoI (5′-CCGCTCGAGAGCTACTGGAACAATGACTCG-3′). The PCR products were digested with NdeI and XhoI enzymes and subsequently cloned into the pET16b vector (Novagen). Freshly transformed BL21(DE3) cells harboring the pET16b::rpoH plasmid were grown in 500 ml of 2× YT medium (64) at 30°C until turbidity at 660 nm reached 0.4. Cultures were then incubated at 16°C and incubated with 0.25 mM IPTG for 3 h, after which cells were pelleted and frozen. The cell pellet was resuspended in 80 ml of lysis buffer (50 mM Tris-HCl, pH 8, 50 mM NaCl, 0.5 mM EDTA, pH 8, 5% [vol/vol] glycerol, 10 mM imidazole, 10 μl of benzonase [Roche], and 1 mM Complete protease inhibitor mixture [Roche Applied Science]) and disrupted in a French pressure cell. After centrifugation at 30,000 × g for 1 h, the soluble extract was passed through a 0.45-μm-pore-size filter and loaded onto a 5-ml Ni-agarose column (Amersham Biosciences) preequilibrated with buffer A-RpoH (50 mM Tris-HCl, pH 8, 50 mM NaCl, 0.5 mM EDTA, pH 8, 5% glycerol, 10 mM imidazole). The column was washed with buffer A until nonspecifically bound material was removed. RpoH eluted from the column at about 400 mM imidazole in a 30-ml imidazole gradient (from 55 mM to 750 mM imidazole) in buffer B-RpoH (buffer A supplemented with 750 mM imidazole). Eluted fractions containing RpoH protein were dialyzed against storage buffer (50 mM Tris-HCl, pH 8, 50 mM NaCl, 0.5 mM EDTA, pH 8, 20% glycerol) and stored at −70°C until use. Freshly transformed BL21(DE3) cells harboring the pET16b::rpoS plasmid were grown in 500 ml of 2× YT medium (64) at 30°C until turbidity at 660 nm reached 0.4. Cultures were then incubated at 16°C and incubated with 0.25 mM IPTG for 3 h, after which cells were pelleted and frozen. The cell pellet was resuspended in 80 ml of lysis buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 0.5 mM EDTA, 2 mM β-mercaptoethanol, 10% [vol/vol] glycerol, 10 mM imidazole, 10 μl of benzonase [Roche] and 1 mM Complete protease inhibitor mixture [Roche Applied Science]) and disrupted in a French pressure cell. After centrifugation at 30,000 × g for 1 h, the soluble extract was passed through a 0.45-μm-pore-size filter and loaded onto a 5-ml Ni-agarose column (Amersham Biosciences) preequilibrated with buffer A-RpoS (20 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM β-mercaptoethanol, 10% glycerol, and 10 mM imidazole). The column was washed with buffer A until nonspecifically bound material was removed. RpoS eluted from the column at about 350 mM imidazole in a 30-ml imidazole gradient in buffer B-RpoS (buffer A supplemented with 1 M imidazole). Eluted fractions containing RpoS protein were dialyzed against storage buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 10% glycerol, 1 mM DTT) and stored at −70°C until use. For both preparations, electrophoresis of 10 μg of protein solution gave a single Coomassie-stained band corresponding to the purified sigma factors.
Single-round in vitro transcription assays.
The template was a linear Pm fragment obtained as a 310-bp PCR product (positions −114 to +195 of the Pm promoter) using oligonucleotides Pm300H (5′-GCCAAGCTTGGGCGAGATAAATCCAGTTGC-3′) and Pm141E (5′-GGAATTCGGCTGCAGTGTCCGGTTTG-3′). We used recombinant RNA polymerase holoenzymes reconstituted by incubation of E. coli core RNA polymerase (Epicenter) with purified P. putida His-tagged σ32 or σ38 sigma factors (1:5 ratio) in transcription buffer L (50 mM Tris-HCl, pH 8, 200 mM potassium glutamate, 10 mM magnesium-acetate, 2 mM DTT, 250 μM ATP, and 100 μg/ml bovine serum albumin [BSA]) for 15 min at 30°C. It is worth noting that P. putida and E. coli RNA polymerases are highly homologous and show more than 75% identity in all subunits, including alternative σ factors. Early work showed that E. coli and P. putida RNA polymerases were equally active on P. putida promoters and contacted the same positions in DNA (18, 44). Linear DNA template (5 nM) and reconstituted RNA polymerase holoenzymes (50 nM) were incubated for 10 min at 30°C in transcription buffer L to allow RNA polymerase-promoter complexes to form. Elongation was started by the addition of a prewarmed mixture containing nucleotides and heparin (final concentrations were 250 μM GTP and CTP, 50 μM UTP, 2 μCi of [α-P32]UTP at 3,000 Ci/mmol, and 100 μg/ml heparin) to the template-polymerase mixture and was allowed to proceed for 10 min at 30°C. Reactions were stopped by the addition of 10 μl of loading buffer (formamide containing 20 mM EDTA, xylene cyanol, and bromophenol blue). Samples were electrophoresed in a 5.5% (wt/vol) polyacrylamide denaturing sequencing gel. The results were analyzed with Molecular Imager FX equipment (Bio-Rad, Madrid, Spain).
In vitro KMnO4 footprinting experiments.
Potassium permanganate footprinting is based on the hyperreactivity of single-stranded thymines to this compound, a reaction that makes it possible to probe open complex formation by RNA polymerase (65). A labeled 158-bp DNA fragment containing the Pm promoter (positions −113 to +35) was generated by PCR using pJLR100 as a template with the primers Pm141E (5′-GGAATTCGGCTGCAGTGTCCGGTTTG-3′) and Pm141H (5′CCCAAGCTTGTCATGGTCATGACTCC-3′). Reconstituted RNA polymerases prepared as above (a 50 nM concentration of E. coli RNA polymerase core from Epicenter) and the Pm promoter fragment were incubated in transcription buffer (40 mM HEPES, pH 8, 100 mM potassium glutamate, 10 mM MgCl2, 2 mM DTT, 100 μg/ml BSA) for 15 min at 37°C in a final volume of 20 μl; when indicated, incubation reaction mixtures also contained a 500 nM concentration of XylS-C (see above). Freshly prepared KMnO4 was added to a final concentration of 10 mM, and the reaction was stopped after 30 s by adding 50 μl of a solution containing 1.5 M β-mercaptoethanol, 3 M ammonium acetate, and 0.1 mM EDTA. The samples were phenol extracted, and glycogen was added to a final concentration of 0.1 mg/ml; samples were precipitated with 100% ethanol, washed with 70% ethanol, and resuspended in 40 μl of piperidine (1 M). After 30 min at 90°C, 80 μl of a solution containing 0.3 M sodium acetate, pH 5.2, and 250 μg/ml glycogen was added, and samples were ethanol precipitated, washed with 70% ethanol, and resuspended in 8 μl of loading buffer. Urea-polyacrylamide sequencing gels were calibrated with Maxam-Gilbert G+A sequencing reactions of the labeled fragment and quantified with a Bio-Rad Molecular Imager FX and Quantity One software.
Chromatin immunoprecipitation) ChIP.
The E. coli CC118λPm::lacZ strain carrying a Pm::lacZ fusion with or without a plasmid bearing the wild-type XylS (pERD103) was grown overnight in LB medium with or without kanamycin (25 μg/ml), respectively, at 30°C and 200 rpm. Flasks containing 25 ml of fresh medium were inoculated with an aliquot fraction of these cultures and incubated at 30°C. When the optical density at 600 nm reached 0.3 to 0.5, 1 mM 3MB was added, and incubation continued for 20 min in the presence or absence of 3MB. Cells were treated in vivo with formaldehyde cross-linking agent (1% final concentration). After 20 min, cross-linking was quenched by the addition of glycine (0.5 M final concentration), and DNA was extracted from lysed cells and sheared by sonication as described previously (19). Monoclonal mouse antibodies against the β-subunit of RNA polymerase were obtained from Neoclone (Madison, WI), and rabbit polyclonal anti-XylS was produced by Eurogentec (EGT Group, Belgium). Immunoprecipitations using antibodies against XylS or subunits of RNA polymerase were carried out as previously described (19), except that antibody-nucleoprotein incubations were done overnight at 4°C, and all subsequent washing steps were performed at 4°C. Before analysis, DNA was purified from the immunoprecipitate with a PCR purification kit (Qiagen) and resuspended in 200 μl of water. After purification, real-time PCR was used to analyze immunoprecipitated DNA; PCR primers mltA1 and mltA2 for mannitol permease were used as the reference to correct for errors in DNA concentration used for each assay, and PCR primers Pm3 and Pm4 were used to amplify the Pm promoter. Real-time PCR was performed on an iCycler iQ detection system according to the manufacturer's instructions. The PCRs (10 μl) were set up with the following reagents: 5 μl of 2× Sybr Green Supermix (Bio-Rad), 2.5 μl of immunoprecipitated DNA samples, and a 1 μM concentration of each oligonucleotide primer. The thermal cycling conditions used were as follows: 10 min at 95°C followed by 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s. A final melt curve was plotted to check the specific amplification of both probes. All reactions were run in triplicate. Quantitative PCR (QT-PCR) results with the specific Pm primers were normalized against QT-PCR results obtained with control mtlA primers for each sample.
β-Galactosidase assays.
E. coli CC118λPm::lacZ cells bearing plasmid pGP1-2 were transformed with pBBR-XylS-C or both pBBR1-XylS-C plus a compatible plasmid bearing XylS-N (pET16b::XylS-N) (Table 1). Transformants were grown overnight on LB medium containing the appropriate antibiotics. Three independent clones of each strain were used. Duplicate cultures were prepared by diluting cells from overnight cultures to 1/100. After 1 h at 30°C, the cultures were supplemented with 0.2 mM IPTG. After 2 h at 30°C, one of the duplicates was induced with 1 mM 3MB. Samples for β-galactosidase activity assays were taken 2 h after induction. β-Galactosidase activity was determined in permeabilized whole cells according to Miller (49).
RESULTS
XylS specifically binds directs repeats in Pm sequence.
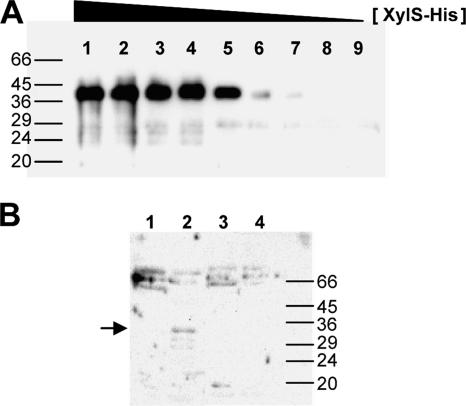
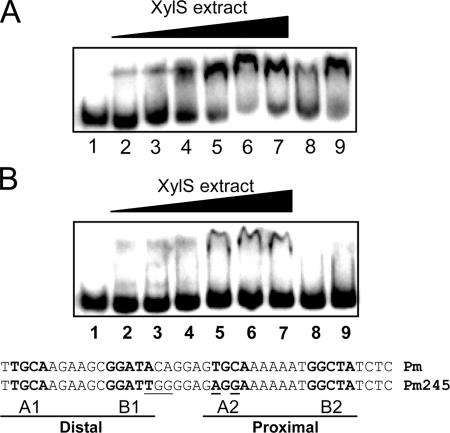
XylS, like most members of the AraC family, aggregates when its concentration exceeds a threshold value, hindering biochemical analysis (2, 28, 48, 50, 73). To overcome this problem, we devised a method to work with XylS-enriched extracts from which most DNA-binding proteins had been removed through heparin chromatography (see Materials and Methods). Briefly, crude extracts prepared from E. coli CC118 cultures expressing XylS protein or mutant derivatives at moderate levels were loaded onto a heparin column. Fractions were analyzed in Western blots with polyclonal anti-XylS antiserum (Fig. 2B). A 36-kDa protein band corresponding to XylS was clearly detected in the heparin unbound fraction. Bound and unbound fractions were tested with EMSAs for their ability to bind a 100-bp DNA fragment containing the Pm regulatory region, spanning positions −10 to −110 with respect to the transcriptional start point. Only the heparin-unbound fraction was able to form a stable DNA-protein complex, which appeared as a clearly shifted band (Fig. 3A). The presence of an excess of unlabeled competitor DNA could titrate XylS from its target DNA (Fig. 3A, lane 8), whereas the addition of nonspecific DNA did not reduce the shifted DNA-protein complex (Fig. 3A, lane 9). To rule out the possibility of Pm-DNA-binding proteins in the extracts, the labeled Pm target was incubated with increasing concentrations of similarly processed extracts either with or without the XylS protein. Only extracts containing XylS produced retardation in EMSAs (data not shown). XylS protein present in the enriched extracts was quantified in Western blots using known amounts of purified XylS as a reference (Fig. 2A). Assuming a cell cytoplasm volume of 6.7 × 10−16 liters (69; http://redpoll.pharmacy.ualberta.ca/CCDB/cgi-bin/STAT_NEW.cgi), we could estimate that XylS concentration in the cell was in the range of 50 nM, which corresponds to about 200 molecules per cell, in agreement with reported SoxS basal levels of 100 molecules per cell (21). It is worth noting that these concentrations are two orders of magnitude lower than the XylS concentration in the extracts used for EMSA (see below).
FIG. 2.
Heparin chromatography of XylS-containing extracts and estimation of XylS concentration in the extracts. (A) XylS was purified from inclusion bodies after solubilization with 6 M guanidium, renaturing, and His-affinity chromatography. It is worth noting that XylS protein obtained with this protocol was inactive. Samples of known XylS concentration (5, 3.5, 2.5, 1.5, 1, 0.5, 0.1, 0.01, and 0.001 μg loaded in lanes 1 to 9, respectively) were separated by denaturing sodium dodecyl sulfate-PAGE, transferred to a nitrocellulose membrane, and probed with antibodies at a dilution of 1/1,000 against XylS (76). (B) Cell extracts (170 μg of total protein) of E. coli CC118 (pLOW2::XylS) (lanes 1 and 2) or E. coli CC118 (pLOW2) (lanes 3 and 4) were loaded in a 1-ml heparin column, and samples were eluted as indicated in Materials and Methods. Lanes 1 and 3, whole extract; lanes 2 and 4, extract eluted from the column. Molecular weight markers were BSA (66 kDa), ovalbumin (45 kDa), pepsin (36 kDa), carbonic anhydrase (29 kDa), trypsinogen (24 kDa), and trypsin inhibitor (20 kDa).
FIG. 3.
DNA binding of XylS to Pm promoter. EMSA for binding of 32P-labeled wild-type Pm DNA fragment (A) or mutant Pm245 DNA fragment (B) by purified extracts containing wild-type XylS protein. EMSA was performed as described in Materials and Methods with either no protein added (lane 1) or increasing amounts (0.5, 1, 2, 5, 10, and 15 μg) of XylS-enriched extracts (lanes 2 to 7). An excess of specific (0.5 μg of wild-type Pm DNA fragment; lane 8) or nonspecific competitor [1 μg of poly(dI-dC) DNA; lane 9] was added to reaction mixtures that also contained 15 μg of crude extract. Wild-type and mutant Pm sequences used in panels A or B, respectively, are shown. A and B box sequences are shown in bold, and mutations in Pm245 nucleotides are underlined.
Pm245 is a mutant promoter with five point mutations in the XylS binding site (Fig. 3B) and accordingly produces no detectable XylS-dependent transcriptional activity in vivo (16). The results shown in Fig. 3B demonstrate that XylS binding to Pm245 mutant DNA was significantly reduced, supporting the notion that the XylS protein present in cell extracts specifically bound Pm in the previous EMSA.
The EMSA experiments gave identical results when 3MB was added to the assays or when the extracts were obtained from cultures grown in the presence of 1 mM 3MB (data not shown), indicating that under these protein concentrations, the wild-type regulator was able to bind the target promoter in vitro in the absence of effectors.
A second role for 3MB in XylS binding to the Pm promoter.
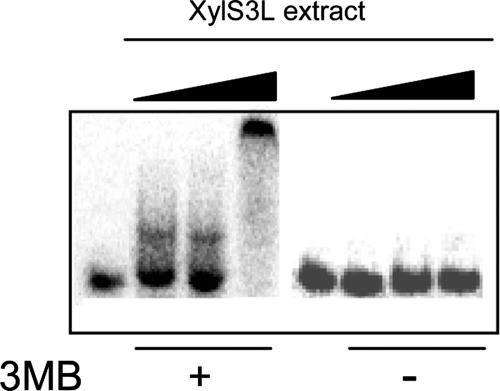
Studies with members of the AraC family suggest that the proteins lacking an N-terminal domain bind DNA as monomers, whereas the majority of two-domain proteins are dimers in solution and most probably bind DNA as dimers (7, 52, 53, 67). It has been suggested that effector binding to the XylS N-terminal moiety causes a conformational change which favors in vivo dimer formation (61). The triple mutant XylS(L193A L194A I205A) [referred to as XylS(3L)], in which alanine replaces three residues located in the antiparallel coiled-coil region connecting the N- and C-terminal domains, was unable to form stable dimers in vivo and was unable to stimulate transcription in either the presence or absence of 3MB (61). To investigate the influence of dimerization on XylS DNA binding and the possible role of 3MB in this process, we prepared cell extracts from E. coli cultures expressing the XylS(3L) mutant. EMSA with extracts containing XylS(3L) in the absence of 3MB did not yield shifted bands, indicating that under these conditions the mutant XylS protein was unable to bind DNA. However, when 3MB was present, two well-defined shifted bands were observed (Fig. 4): at low XylS(3L) concentrations a rapidly migrating band was observed; a fainter band with the same mobility was sometimes observed in EMSA using very low concentrations of wild-type XylS. A second, more retarded band appeared at higher protein concentrations, coincident with the migration band observed with wild-type XylS (not shown).
FIG. 4.
DNA binding of mutant XylS(3L) to wild-type Pm promoter. EMSA for binding of 32P-labeled wild-type Pm DNA fragment in the presence (+) or absence (−) of 3MB by purified extracts containing mutant XylS(3L) protein. Mobility shift assays were performed as described in Materials and Methods with either no protein added (first and fifth lanes from the left) or increasing amounts (0.5, 1, and 10 μg) of extracts that contained XylS(3L) (second to fourth lane and sixth to eighth lane). Controls with an excess of nonlabeled Pm DNA strongly reduced the amount of shifted DNA, while the addition of excess unspecific DNA did not modify the shift.
The fact that at high concentrations (approximately a 5 μM concentration of protein in XylS-enriched extracts, as estimated in Western blots) (Fig. 2), wild-type XylS was able to bind the Pm promoter in the absence of 3MB (Fig. 3A) is consistent with the 3MB-independent activation observed in vivo when XylS is overproduced (45, 46, 56). Thus, high XylS protein concentrations favor dimer formation, as previously shown by Ruíz et al. (61) in cross-linking experiments. This is also in accordance with previous results showing that dimer formation was 3MB independent in vitro, although it required 3MB in vivo when XylS protein was produced at low levels (61). Taken together, the results with wild-type and mutant XylS(3L) proteins in vivo and in vitro provide evidence that DNA binding, which can be obtained either with a XylS dimer irrespective of the presence of 3MB or with a protein allele unable to dimerize in the presence of the effector, and protein dimerization are two connected but independent processes. The dependence of XylS(3L) DNA binding on the presence of 3MB points to the need of a conformational change in this protein. However, the effect of 3MB on the wild-type protein DNA binding could only be determined in vivo at low XylS concentrations (see below).
The XylS-N inhibits C-terminal domain activity.
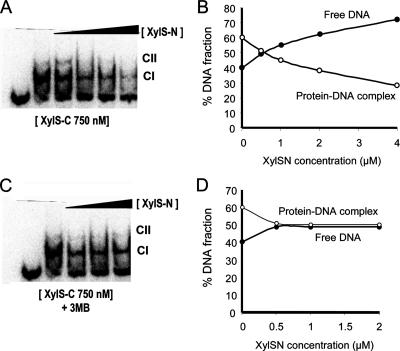
XylS-C, lacking the N-terminal domain, shows constitutive activity, i.e., it activates the Pm promoter in the absence of effector (Domínguez-Cuevas et al., unpublished). This suggests an inhibitory effect of the N-terminal domain on XylS-C binding activity (32). If this were the case, we predict that the two domains should be able to interact in solution; this interaction would negatively influence XylS-C DNA binding, and 3MB would modulate this interaction. To test this hypothesis, we purified both the N- and C-terminal domains of XylS using established protocols (see Materials and Methods; Domínguez-Cuevas et al., unpublished), and we used EMSA to analyze XylS-C-DNA complex formation. A fixed concentration of XylS-C (500 nM) was preincubated with increasing amounts of purified XylS-N (from 500 nM to 4 μM) prior to the addition of labeled Pm promoter probe. Quantification of the complexes formed in each assay showed that the presence of XylS-N in the mixtures repressed XylS-C binding to DNA, suggesting that the two domains established direct protein-protein interactions even when they were not connected by a linker (Fig. 5). When 3MB was added to the preincubation mixture, the inhibition exerted by the N-terminal domain was significantly reduced (Fig. 5). To confirm the relevance of this intramolecular inhibition in XylS, we analyzed the ability of the N-terminal domain to repress C-terminal domain activation of Pm in vivo. To that end, we cloned the N- and C-terminal domains in compatible plasmids under the control of a hybrid T7-Plac promoter (pET16b::XylS-N) and a Plac promoter (pBBR1::XylS-C). Strain CC118λPm::lacZ (pGP1-2), which bears a Pm::lacZ fusion in the chromosome and a plasmid expressing the T7 polymerase (70), was transformed with pBBR1::XylS-C or with both pBBR1::XylS-C and pET16b::XylS-N. Pm-dependent β-galactosidase activity was measured in exponentially growing IPTG-induced cells in the presence or absence of 3MB. Table 2 shows that in the absence of effector, XylS-C induced Pm transcription, and this activity was repressed 8.3 times in the presence of XylS-N. However, in the presence of 3MB, the repression exerted by XylS-N was partially released, thus confirming the observations with purified domains. These results suggest that the N-terminal region interacts directly with the C-terminal domain, preventing its ability to bind DNA. The effector 3MB acts to release repression, and this can be visualized as a direct interaction triggering a crucial conformational change, which simultaneously releases the protein constraints on both dimerization and DNA binding.
FIG. 5.
N-terminal domain repression of XylS-C binding to Pm DNA. EMSA for binding of 32P-labeled wild-type Pm DNA fragment by XylS-C in the presence and absence of increasing concentrations of purified XylS-N in the absence (A and B) or presence (C and D) of 1 mM 3MB. EMSAs were performed as described in Materials and Methods with either no protein added or a fixed amount of purified XylS-C (750 nM) in the presence of increasing concentrations of XylS-N (from 0.5 to 4 μM). (B and D) The fraction of total radiolabel in each band from each lane was quantified and plotted as a function of XylS-N concentration: black circles, free DNA; white circles, XylS-C-DNA complex.
TABLE 2.
In vivo N-terminal domain repression of C-terminal domain-dependent Pm activation
| Proteins present in the tested strain
|
β-Gal activityb
|
|||
|---|---|---|---|---|
| XylS-C | XylS-N | T7 RNAPa | Without 3MB | With 3MB |
| + | − | + | 100 ± 9 | 105 ± 7 |
| + | + | + | 12 ± 2 | 24 ± 3 |
RNAP, RNA polymerase.
β-Galactosidase activity, measured in relative units (100% activity corresponded to 156 Miller units). Xyls-N repressed activity 8.3-fold in the absence of the 3MB effector but only 4.4-fold in the presence of 3MB.
3MB activation of XylS results in RNA polymerase recruitment to Pm.
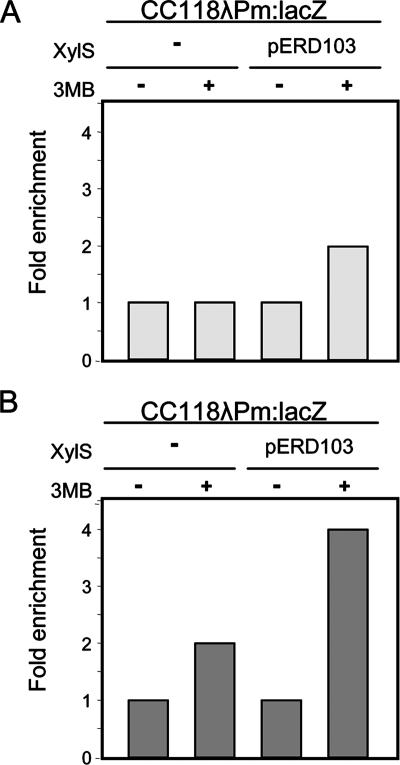
XylS, which activates transcription from the Pm promoter in response to 3MB, is suggested to establish direct interactions with the RNA polymerase α-subunit carboxy-terminal domain(63). In addition, the 2-bp overlap between the proximal XylS site and the −35 hexamer element suggests direct contacts between XylS and the σ factor, as has been shown for other members of the family (20, 78). To investigate the steps leading to this interaction and the influence of 3MB, we measured XylS and RNA polymerase binding to the Pm promoter in the presence or absence of 3MB using ChIP. E. coli strain CC118λPm::lacZ carrying a Pm::lacZ fusion with or without a plasmid bearing the wild-type XylS (pERD103) was grown in the presence or absence of 3MB. Cells were treated with formaldehyde to freeze in vivo protein-DNA interactions, and nucleoprotein was extracted from lysed cells and sheared by sonication. Either polyclonal anti-XylS antiserum or monoclonal anti-β-subunit antibody was used to immunoprecipitate DNA fragments. Figure 6 shows the enrichment levels relative to the control strain CC118λPm::lacZ obtained in real-time PCR analysis with primers specific for the Pm promoter. A pair of primers designed to amplify the mltA promoter was used as a reference (see Materials and Methods). ChIP with XylS antibody resulted in a twofold enrichment of XylS-bound Pm in response to 3MB (Fig. 6A), indicating that the ligand promoted XylS binding to DNA. When immunoprecipitates were prepared with antibodies directed against the RNA polymerase β-subunit, enrichment of the Pm promoter DNA was greatest in cells grown in the presence of both XylS activator and 3MB (Fig. 6B). This observation parallels the previous finding of MelR recruiting RNA polymerase to the target PmelAB promoter at a similar level (19) and supports the hypothesis that XylS enhances the association of RNA polymerase to the Pm promoter in the presence of 3MB; in other words, XylS recruits RNA polymerase to Pm. Figure 6B also shows that RNA polymerase also bound the Pm promoter in response to 3MB even in the absence of the XylS activator, consistent with the previously observed increase in Pm transcription during the stress response (39), such as the one triggered by 3MB.
FIG. 6.
ChIP analysis of RNA polymerase and XylS binding to the Pm promoter. ChIP was carried out as described in Materials and Methods. XylS binding to the Pm promoter in the presence and absence of 3MB was determined. E. coli CC118λPm::lacZ with or without the plasmid pERD103 was grown in the presence or absence of 3MB. Data show the ratio of real-time PCR quantitation of anti-XylS (A) or anti-β-subunit (B) antibody immunoprecipitate to nonspecific precipitate without antibody, generated by the Pm promoter sequence and corrected with reference to the mlt sequence, in each strain and under each growth condition. Data represent the enrichment of QT-PCR product relative to the control CC118λPm::lacZ strain in the absence of 3MB. Results are the average of three independent experiments.
XylS promotes open complex formation and transcription initiation at the Pm promoter.
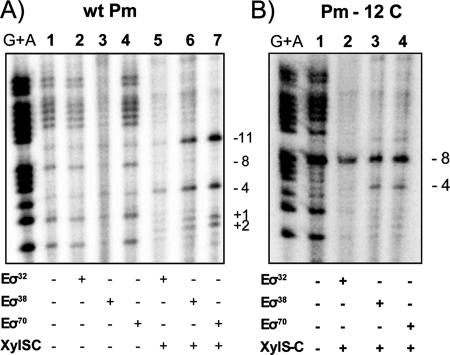
Previous studies showed that the Pm promoter could be served in vivo by two different RNA polymerases: Eσ32 in exponential phase and Eσ38 in stationary phase (9, 39). To confirm this unusual σ requirement, we used KMnO4 footprinting and analyzed open complex formation mediated by Eσ32, Eσ38, and Eσ70. RNA polymerase holoenzyme with the alternative sigma factors σ32 and σ38 was reconstituted with commercial E. coli core and purified P. putida σ32 or σ38 factors (see Materials and Methods). An end-labeled DNA fragment containing Pm was treated with potassium permanganate after preincubation with each of the three different reconstituted RNA polymerases to assess if complex formation depended on the presence of XylS protein. The assays were performed in the presence or in the absence of XylS-C, the truncated version of XylS devoid of the effector recognition domain (see Materials and Methods). This protein reproduces XylS features except that it does not require 3MB to bind DNA or activate transcription (Domínguez-Cuevas et al., unpublished). Figure 7 shows that, in the absence of XylS-C, none of the reconstituted alternative RNA polymerases produced significant changes in the reactivity pattern with respect to controls (Fig. 7A, lanes 2 to 4). However, in the presence of XylS-C, all three RNA polymerases induced clear reactivity of the thymines between positions −11 and +2 of the Pm promoter (Fig. 7A, lanes 5 to 7). It is known that RNA polymerase holoenzymes containing either σ38 or σ70 recognize the nontemplate strand in the −10 region, where T at position −11 plays a central role (41, 60). The reactivity pattern was identical with each of the three RNA polymerases, except that intensity was lower with Eσ32. Although the involvement of σ32 in transcription from Pm is well documented in vivo (39), we did not obtain a stronger signal in vitro with our experimental conditions. If only commercial E. coli RNA polymerase core was used in the assays to test for the presence of any remnant σ factor, the pattern was similar to the controls without XylS. When we repeated the experiments with the Pm-12C mutant promoter (described as severely defective in transcription [16]), a hyperactive band at −8, not observed with wild-type Pm, was clearly visible with all three RNA polymerases (Fig. 7B). The reactivity pattern for all three RNA polymerases was different from the wild-type Pm pattern and, as explained above, unlikely to be due to a productive transcription bubble formation. It is worth noting that this is the first biochemical evidence of two alternative RNA polymerases recognizing the Pm promoter at the same initiation site, both in a XylS-dependent manner, and confirms previous results obtained in vivo (13, 39).
FIG. 7.
Potassium permanganate footprinting of the Pm promoter. The DNA (bottom strand) in the presence of either no protein (lane 1) or RNA polymerase with different sigma factors in the presence (lanes 5 to 7 in A and lanes 2 to 4 in B) or absence (lanes 2 to 4 in A) of XylS-C was modified with potassium permanganate (10 mM) and cleaved with piperidine. The numbers (−11, −8, −4, +1, and +2) indicate the cleavage sites. Wt, wild type.
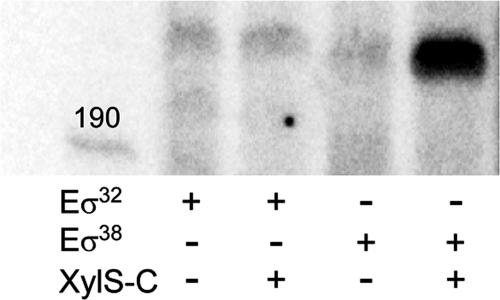
To confirm the role of XylS in Pm transcription, we explored its ability to activate transcription in single-round transcription assays with a linear DNA template carrying the Pm wild-type promoter between positions −113 and +35. Reactions were carried out in the presence or absence of XylS. Because of the tendency of XylS to aggregate, XylS-C, a soluble constitutive truncated XylS derivative, was used (Domínguez-Cuevas et al., unpublished). Results in Fig. 8 show a strong signal corresponding to the expected transcription product with Eσ38, which was considerably weaker with Eσ32, though detectable. In addition, transcription with Eσ38 was stimulated in the presence of the XylS-C regulator, an effect which was barely detectable with Eσ32. Controls with only E. coli RNA polymerase core in the presence of XylS gave negative results (data not shown).
FIG. 8.
Effect of XylS on Pm transcription. Transcription was performed using reconstituted RNA polymerase with either σ32 or σ38 and XylS-C in the presence of [α-32P]UTP-labeled nucleotide. RNA products were resolved by urea-PAGE. The template was a linear DNA fragment containing Pm and was obtained as indicated in Materials and Methods.
DISCUSSION
The nature of XylS functioning in its cell host requires that this regulator activate transcription from the Pm promoter in response to 3MB and that its overproduction through a toluene-induced cascade regulatory circuit lead to transcriptional activation in a 3MB-independent manner (40, 56, 58). Our findings suggest an explanation for this apparent contradiction, and we present evidence supporting a molecular model to explain the mechanism of XylS activation when produced at low and high concentrations.
Activation mechanisms used by AraC family members have been studied in detail in some proteins, and processes such as interaction with RNA polymerase α and σ subunits, N-terminal domain modulation, or DNA looping or overproduction have been implicated in transcriptional activation (23, 30, 37, 43). XylS presents a unique feature not shared by most two-domain AraC proteins: in the absence of effector it is able to reach an active conformational state when its concentration exceeds a threshold value (56). Interestingly, this behavior is similar to the activation mechanism of single-domain proteins in the AraC family, such as MarA and SoxS, where induction of the pathway in response to specific signals increases intracellular levels of the regulator and triggers activation of the target promoter (22, 23). XylS shares with MarA and Rob the feature that they all respond to signal molecules that induce the stress response; in fact, both 3MB (the effector of XylS) and toluene (which triggers the cascade circuit leading to XylS overproduction) can induce the heat-shock response (8, 39, 75). Furthermore, XylS activation of the Pm promoter was functional with Eσ32 RNA polymerase (Fig. 7 and 8), consistent with the requirement of this RNA polymerase for transcription in vivo (39). These observations point to a stress response strategy based on regulator overproduction. The evolutionary implications of this possibility are interesting: considering the distinct two-domain structure of XylS among the stress-responsive AraC family regulators, one is tempted to suggest that XylS may have originated from a fusion protein that joined two functional domains, a MarA-like DNA-binding element and an aromatic responsive element. In fact, the XylS C-terminal domain can activate transcription constitutively (unpublished), and this was also been recently shown for RhaS (79). Interestingly, in E. coli an untranslated sequence with homology to the XylS-N sequence was found directly upstream from marA (24). This untranslated sequence was superimposed on the unrelated marR gene although in a different frame.
To address the molecular function of 3MB in Pm activation by XylS, we reasoned that 3MB could activate transcription in two possible ways: (i) directly favoring XylS DNA binding or (ii) promoting protein dimerization, thus indirectly favoring DNA binding. In addition, we examined the mechanism of XylS activation in the absence of 3MB, a physiological process known as “cascade circuit,” which depends on increased XylS concentrations in the cell (12, 56). Many dimeric regulators are in equilibrium between a monomeric and a dimeric conformation in the cell. This implies that increasing the protein concentration would shift the equilibrium toward dimerization and favor DNA binding (54). Examples of this situation are the cascade-overproduced XylS or the high XylS concentrations used in vitro. However, the low basal XylS levels normally found in the cell (50 nM as estimated) require 3MB to shift the equilibrium toward dimerization (61). 3MB is also required to increase XylS affinity for DNA, as shown in Fig. 6B. Thus, we suggest a double role for 3MB in XylS activation under physiological conditions. On the one hand, 3MB-activated conformation would favor and stabilize dimerization, as has previously been shown (61). But in addition the 3MB-induced change would be required to make the DNA binding surface accessible (Fig. 5), a change which could also be achieved through protein dimerization. In this work we have shown that when dimerization was prevented as in the XylS(3L) mutant (61), binding of 3MB to the regulator led to a conformational change essential for DNA binding. This observation, together with the fact that the XylS C-terminal domain was able to activate Pm in the absence of 3MB (P. Domínguez-Cuevas, unpublished data) indicates that the N-terminal domain acts as an intramolecular repressor, which is the result of a direct interaction between independently functional domains and not of the connection of these domains by the flexible linker. Intramolecular repression of the DNA binding domain has been described for members of the NtrC family of regulators such as DmpR and XylR, in which deletion of the N-terminal effector binding domain resulted in an activator that mediated transcription constitutively (11, 51). A similar effector-responsive interaction between the two domains has also been described in AraC (10).
The process of Pm transcription activation requires XylS binding to its operator sequences and binding of RNA polymerase to its specific recognition sequence in the promoter. Further sequential events involve isomerization of the promoter from a closed to an open complex and transcription initiation. We previously showed that transcription from the Pm promoter is mediated in vivo by two alternative RNA polymerases: Eσ32 or Eσ38 (9, 39). We have confirmed that these RNA polymerases are operative at Pm in permanganate footprinting assays of the XylS DNA binding domain, which showed a similar pattern in the presence of each of the three RNA polymerases when the XylS activator was present (Fig. 7). This indicates that the interaction between XylS and RNA polymerase stimulates isomerization from a closed to an open complex, although we cannot determine in these assays whether open complex isomerization is a direct effect of XylS or whether it is a consequence of XylS recruitment of RNA polymerase to Pm. This was further confirmed in in vitro assays showing a clear XylS-C requirement to obtain full transcription activity with σ38.
Previous findings showed that in an rpoS rpoH double mutant, Pm transcription remained at basal levels, suggesting that Eσ70 in vivo activity was low at this promoter (39). This apparent contradiction with the high rate of open complex formation found in vitro with this holoenzyme (Fig. 7A, lane 7) has previously been reported for several promoters, which may be transcribed in vitro by both Eσ38 and Eσ70 but which in vivo are strictly dependent on Eσ38 (71).
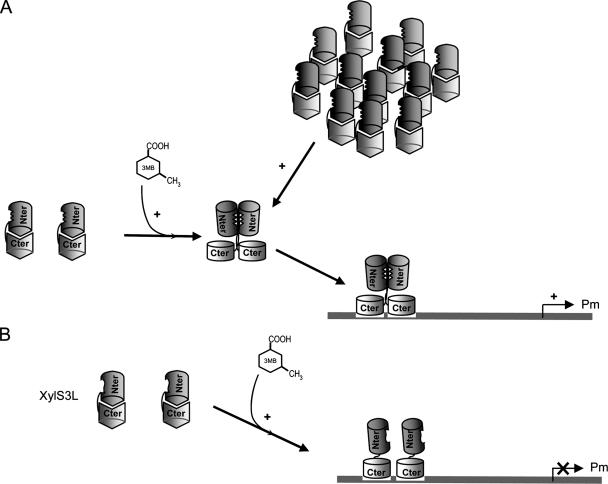
Based on previous and current results, we propose a model to explain the molecular role of 3MB in Pm activation by XylS (Fig. 9). Two processes are central to 3MB functioning. First, 3MB favors XylS DNA binding by altering direct interactions between XylS N-terminal and C-terminal domains (Fig. 5). Second, the suggested conformational change concomitant to dimerization, which occurs either after 3MB activation or through an increase in XylS concentration (61), unmasks the DNA binding domain from the constraint imposed by the N-terminal domain. Dimer formation, also favored at high protein concentrations (61), allows XylS to bind DNA and activate transcription from Pm in the absence of 3MB (Fig. 3). It is worth noting that although 3MB allowed XylS(3L) to bind DNA, this mutant remained unable to activate transcription, showing that XylS dimerization is essential to activate transcription from Pm. Finally, our results show that XylS recruits RNA polymerase to the Pm promoter in response to 3MB and subsequently increases the rate of isomerization of RNA polymerase from closed to open complexes.
FIG. 9.
Model for XylS activation of the Pm promoter. (A) Under basal conditions, XylS DNA-binding domains (light gray barrels) are unable to make contact with DNA. The addition of 3MB produces a conformational change with two important consequences: (i) dimerization interactions are favored between monomers, probably because monomer dimerization regions are exposed; and (ii) the regulator DNA-binding domains are opened, which favors contacts with DNA. Under physiological conditions (i.e., in vivo), XylS does not dimerize unless 3MB is present. When overexpressed or in purified preparations, high concentrations of XylS favor dimerization in the absence of effector. Dimerization might also lead to conformational changes which make DNA-binding domains more available for interactions with target DNA sites. In both cases, XylS binding to Pm in the presence of RNA polymerase activates the promoter. (B) In XylS(3L), 3MB generates the corresponding conformational change, but the result in this case is only the opening of DNA-binding sites, since this mutant is unable to dimerize. Accordingly, the bound protein is not able to promote transcription.
Acknowledgments
This work was supported by MEC projects BCM2001-0515 and BIO2006-05668 and SYSMO EU project GEN 2006-27750-C5-J-E/SYS. P.D.C. is the recipient of a Junta de Andalucía Predoctoral Fellowship in Spain and an EMBO Short Term Fellowship (work at S. Busby's laboratory in Birmingham, United Kingdom).
We thank David Grainger for help with the ChIP experiments and Victor de Lorenzo for providing an efficient anti-XylS antiserum.
Footnotes
Published ahead of print on 22 February 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Wiley, New York, NY.
- 2.Bhende, P. M., and S. M. Egan. 1999. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J. Bacteriol. 1815185-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhende, P. M., and S. M. Egan. 2000. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J. Bacteriol. 1824959-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Busby, S., and R. H. Ebright. 1997. Transcription activation at class II CAP-dependent promoters. Mol. Microbiol. 23853-859. [DOI] [PubMed] [Google Scholar]
- 6.Bustos, S. A., and R. F. Schleif. 1993. Functional domains of the AraC protein. Proc. Natl. Acad. Sci. USA 905638-5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caswell, R., C. Webster, and S. Busby. 1992. Studies on the binding of the Escherichia coli MelR transcription activator protein to operator sequences at the MelAB promoter. Biochem. J. 287501-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domínguez-Cuevas, P., J. E. González-Pastor, S. Marqués, J. L. Ramos, and V. de Lorenzo. 2006. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J. Biol. Chem. 28111981-11991. [DOI] [PubMed] [Google Scholar]
- 9.Domínguez-Cuevas, P., P. Marín, J. L. Ramos, and S. Marqués. 2005. RNA polymerase holoenzymes can share a single transcription start site for the Pm promoter. Critical nucleotides in the −7 to −18 region are needed to select between RNA polymerase with σ38 or σ32. J. Biol. Chem. 28041315-41323. [DOI] [PubMed] [Google Scholar]
- 10.Eustance, R. J., and R. F. Schleif. 1996. In vivo association of protein fragments giving active AraC. Proteins 25501-505. [DOI] [PubMed] [Google Scholar]
- 11.Fernández, S., V. de Lorenzo, and J. Pérez-Martín. 1995. Activation of the transcriptional regulator XylR of Pseudomonas putida by release of repression between functional domains. Mol. Microbiol. 16205-213. [DOI] [PubMed] [Google Scholar]
- 12.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs from a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the compound used for growth. J. Bacteriol. 1782356-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallegos, M. T., S. Marqués, and J. L. Ramos. 1996. The TACAN4TGCA motif upstream from the −35 region in the σ70-σS-dependent Pm promoter of the TOL plasmid is the minimum DNA segment required for transcription stimulation by XylS regulators. J. Bacteriol. 1786427-6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol Rev. 61393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Pérez, M. M., S. Marqués, P. Domínguez-Cuevas, and J. L. Ramos. 2002. XylS activator and RNA polymerase binding sites at the Pm promoter overlap. FEBS Lett. 519117-122. [DOI] [PubMed] [Google Scholar]
- 16.González-Pérez, M. M., J. L. Ramos, M. T. Gallegos, and S. Marqués. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 2742286-2290. [DOI] [PubMed] [Google Scholar]
- 17.González-Pérez, M. M., J. L. Ramos, and S. Marqués. 2004. Cellular XylS levels are a function of transcription of xylS from two independent promoters and the differential efficiency of translation of the two mRNAs. J. Bacteriol. 1861898-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gragerov, A. I., A. A. Chenchik, V. A. Aivasashvilli, R. Beabealashvilli, and V. G. Nikiforov. 1984. Escherichia coli and Pseudomonas putida RNA polymerases display identical contacts with promoters. Mol. Gen. Genet. 195511-515. [DOI] [PubMed] [Google Scholar]
- 19.Grainger, D. C., T. W. Overton, N. Reppas, J. T. Wade, E. Tamai, J. L. Hobman, C. Constantinidou, K. Struhl, G. Church, and S. J. Busby. 2004. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J. Bacteriol. 1866938-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grainger, D. C., C. L. Webster, T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase sigma subunit. Mol. Microbiol. 511297-1309. [DOI] [PubMed] [Google Scholar]
- 21.Griffith, K. L., I. M. Shah, T. E. Myers, M. C. O'Neill, and R. E. Wolf, Jr. 2002. Evidence for “pre-recruitment” as a new mechanism of transcription activation in Escherichia coli: the large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem. Biophys. Res. Commun. 291979-986. [DOI] [PubMed] [Google Scholar]
- 22.Griffith, K. L., I. M. Shah, and R. E. Wolf, Jr. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 511801-1816. [DOI] [PubMed] [Google Scholar]
- 23.Griffith, K. L., and R. E. Wolf, Jr. 2004. Genetic evidence for pre-recruitment as the mechanism of transcription activation by SoxS of Escherichia coli: the dominance of DNA binding mutations of SoxS. J. Mol. Biol. 3441-10. [DOI] [PubMed] [Google Scholar]
- 24.Hachler, H., S. P. Cohen, and S. B. Levy. 1996. Untranslated sequence upstream of MarA in the multiple antibiotic resistance locus of Escherichia coli is related to the effector-binding domain of the XylS transcriptional activator. J. Mol. Evol. 42409-413. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, L. H., S. J. Sorensen, and L. B. Jensen. 1997. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186167-173. [DOI] [PubMed] [Google Scholar]
- 27.Holcroft, C. C., and S. M. Egan. 2000. Interdependence of activation at rhaSR by cyclic AMP receptor protein, the RNA polymerase alpha subunit C-terminal domain, and RhaR. J. Bacteriol. 1826774-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jair, K. W., R. G. Martin, J. L. Rosner, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1995. Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J. Bacteriol. 1777100-7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jair, K. W., X. Yu, K. Skarstad, B. Thony, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J. Bacteriol. 1782507-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahramanoglou, C., C. L. Webster, M. S. El-Robh, T. A. Belyaeva, and S. J. Busby. 2006. Mutational analysis of the Escherichia coli melR gene suggests a two-state concerted model to explain transcriptional activation and repression in the melibiose operon. J. Bacteriol. 1883199-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaldalu, N., T. Mandel, and M. Ustav. 1996. TOL plasmid transcription factor XylS binds specifically to the Pm operator sequence. Mol. Microbiol. 20569-579. [DOI] [PubMed] [Google Scholar]
- 32.Kaldalu, N., U. Toots, V. de Lorenzo, and M. Ustav. 2000. Functional domains of the TOL plasmid transcription factor XylS. J. Bacteriol. 1821118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kessler, B., M. Herrero, K. N. Timmis, and V. de Lorenzo. 1994. Genetic evidence that the XylS regulator of the Pseudomonas TOL meta operon controls the Pm promoter through weak DNA-protein interactions. J. Bacteriol. 1763171-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol 7424-430. [DOI] [PubMed] [Google Scholar]
- 35.Landini, P., and S. J. Busby. 1999. The Escherichia coli Ada protein can interact with two distinct determinants in the σ70 subunit of RNA polymerase according to promoter architecture: identification of the target of Ada activation at the alkA promoter. J. Bacteriol. 1811524-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landini, P., T. Gaal, W. Ross, and M. R. Volkert. 1997. The RNA polymerase alpha subunit carboxyl-terminal domain is required for both basal and activated transcription from the alkA promoter. J. Biol. Chem. 27215914-15919. [DOI] [PubMed] [Google Scholar]
- 37.Lobell, R. B., and R. F. Schleif. 1990. DNA looping and unlooping by AraC protein. Science 250528-532. [DOI] [PubMed] [Google Scholar]
- 38.Manzanera, M., S. Marqués, and J. L. Ramos. 2000. Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett. 476312-317. [DOI] [PubMed] [Google Scholar]
- 39.Marqués, S., M. Manzanera, M. M. González-Pérez, M. T. Gallegos, and J. L. Ramos. 1999. The XylS-dependent Pm promoter is transcribed in vivo by RNA polymerase with sigma 32 or sigma 38 depending on the growth phase. Mol. Microbiol. 311105-1113. [DOI] [PubMed] [Google Scholar]
- 40.Marqués, S., and J. L. Ramos. 1993. Transcriptional control of the Pseudomonas putida TOL plasmid catabolic pathways. Mol. Microbiol. 9923-929. [DOI] [PubMed] [Google Scholar]
- 41.Marr, M. T., and J. W. Roberts. 1997. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science 2761258-1260. [DOI] [PubMed] [Google Scholar]
- 42.Martin, K., L. Huo, and R. F. Schleif. 1986. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc. Natl. Acad. Sci. USA 833654-3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4132-137. [DOI] [PubMed] [Google Scholar]
- 44.Mermod, N. 1986. Étude de l'expresion de gènes cataboliques chez Pseudomonas. Ph.D. thesis. University of Geneva, Geneva, Switzerland.
- 45.Mermod, N., J. L. Ramos, A. Bairoch, and K. N. Timmis. 1987. The xylS gene positive regulator of TOL plasmid pWWO: identification, sequence analysis and overproduction leading to constitutive expression of meta cleavage operon. Mol. Gen. Genet. 207349-354. [DOI] [PubMed] [Google Scholar]
- 46.Michán, C., B. Kessler, V. de Lorenzo, K. N. Timmis, and J. L. Ramos. 1992. XylS domain interactions can be deduced from intraallelic dominance in double mutants of Pseudomonas putida. Mol. Gen. Genet. 235406-412. [DOI] [PubMed] [Google Scholar]
- 47.Michán, C., L. Zhou, M. T. Gallegos, K. N. Timmis, and J. L. Ramos. 1992. Identification of critical amino-terminal regions of XylS. The positive regulator encoded by the TOL plasmid. J. Biol. Chem. 26722897-22901. [PubMed] [Google Scholar]
- 48.Michán, C. M., S. J. Busby, and E. I. Hyde. 1995. The Escherichia coli MelR transcription activator: production of a stable fragment containing the DNA-binding domain. Nucleic Acids Res. 231518-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 50.Munson, G. P., and J. R. Scott. 1999. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 1812110-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng, L. C., E. O'Neill, and V. Shingler. 1996. Genetic evidence for interdomain regulation of the phenol-responsive final σ54-dependent activator DmpR. J. Biol. Chem. 27117281-17286. [DOI] [PubMed] [Google Scholar]
- 52.Poore, C. A., C. Coker, J. D. Dattelbaum, and H. L. Mobley. 2001. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J. Bacteriol. 1834526-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prouty, M. G., C. R. Osorio, and K. E. Klose. 2005. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol. Microbiol. 581143-1156. [DOI] [PubMed] [Google Scholar]
- 54.Raibaud, O., and M. Schwartz. 1984. Positive control of transcription initiation in bacteria. Annu. Rev. Genet. 18173-206. [DOI] [PubMed] [Google Scholar]
- 55.Ramos, J. L., S. Marqués, and K. N. Timmis. 1997. Transcriptional control of the Pseudomonas TOL plasmid catabolic operons is achieved through an interplay of host factors and plasmid-encoded regulators. Annu. Rev. Microbiol. 51341-373. [DOI] [PubMed] [Google Scholar]
- 56.Ramos, J. L., N. Mermod, and K. N. Timmis. 1987. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol. Microbiol. 1293-300. [DOI] [PubMed] [Google Scholar]
- 57.Ramos, J. L., F. Rojo, L. Zhou, and K. N. Timmis. 1990. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucleic Acids Res. 182149-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramos, J. L., A. Stolz, W. Reineke, and K. N. Timmis. 1986. Altered effector specificities in regulators of gene expression: TOL plasmid xylS mutants and their use to engineer expansion of the range of aromatics degraded by bacteria. Proc. Natl. Acad. Sci. USA 838467-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. USA 9510413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roberts, C. W., and J. W. Roberts. 1996. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell 86495-501. [DOI] [PubMed] [Google Scholar]
- 61.Ruíz, R., S. Marqués, and J. L. Ramos. 2003. Leucines 193 and 194 at the N-terminal domain of the XylS protein, the positive transcriptional regulator of the TOL meta-cleavage pathway, are involved in dimerization. J. Bacteriol. 1853036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruíz, R., and J. L. Ramos. 2002. Residues 137 and 153 at the N terminus of the XylS protein influence the effector profile of this transcriptional regulator and the sigma factor used by RNA polymerase to stimulate transcription from its cognate promoter. J. Biol. Chem. 2777282-7286. [DOI] [PubMed] [Google Scholar]
- 63.Ruíz, R., J. L. Ramos, and S. Egan. 2001. Interactions of the XylS regulators with the C-terminal domain of the RNA polymerase alpha subunit influence the expression level from the cognate Pm promoter. FEBS Lett. 491207-211. [DOI] [PubMed] [Google Scholar]
- 64.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold spring Harbor Laboratory Press, Cold spring Harbor Laboratory, NY.
- 65.Sasse-Dwight, S., and J. D. Gralla. 1989. KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem. 2648074-8081. [PubMed] [Google Scholar]
- 66.Shah, I. M., and R. E. Wolf, Jr. 2004. Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase alpha subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promoters during oxidative stress. J. Mol. Biol. 343513-532. [DOI] [PubMed] [Google Scholar]
- 67.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. The 1.6 Å crystal structure of the AraC sugar-binding and dimerization domain complexed with d-fucose. J. Mol. Biol. 273226-237. [DOI] [PubMed] [Google Scholar]
- 68.Soisson, S. M., B. MacDougall-Shackleton, R. Schleif, and C. Wolberger. 1997. Structural basis for ligand-regulated oligomerization of AraC. Science 276421-425. [DOI] [PubMed] [Google Scholar]
- 69.Sundararaj, S., A. Guo, B. Habibi-Nazhad, M. Rouani, P. Stothard, M. Ellison, and D. S. Wishart. 2004. The CyberCell Database (CCDB): a comprehensive, self-updating, relational database to coordinate and facilitate in silico modeling of Escherichia coli. Nucleic Acids Res. 32D293-D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 821074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanaka, K., Y. Takayanagi, N. Fujita, A. Ishihama, and H. Takahashi. 1993. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc. Natl. Acad. Sci. USA 908303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Terán, W., A. Felipe, A. Segura, A. Rojas, J. L. Ramos, and M. T. Gallegos. 2003. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E TtgABC efflux pump is mediated by the drug binding repressor TtgR. Antimicrob. Agents Chemother. 473067-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Timmes, A., M. Rodgers, and R. Schleif. 2004. Biochemical and physiological properties of the DNA binding domain of AraC protein. J. Mol. Biol. 340731-738. [DOI] [PubMed] [Google Scholar]
- 74.Tobes, R., and J. L. Ramos. 2002. AraC-XylS database: a family of positive transcriptional regulators in bacteria. Nucleic Acids Res. 30318-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Dyk, T. K., T. R. Reed, A. C. Vollmer, and R. A. LaRossa. 1995. Synergistic induction of the heat shock response in Escherichia coli by simultaneous treatment with chemical inducers. J. Bacteriol. 1776001-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Velazquez, F., V. Parro, and V. de Lorenzo. 2005. Inferring the genetic network of m-xylene metabolism through expression profiling of the xyl genes of Pseudomonas putida mt-2. Mol. Microbiol. 571557-1569. [DOI] [PubMed] [Google Scholar]
- 77.Wade, J. T., T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2000. Repression of the Escherichia coli melR promoter by MelR: evidence that efficient repression requires the formation of a repression loop. Mol. Microbiol. 36223-229. [DOI] [PubMed] [Google Scholar]
- 78.Wickstrum, J. R., and S. M. Egan. 2004. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J. Bacteriol. 1866277-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wickstrum, J. R., J. M. Skredenske, A. Kolin, D. J. Jin, J. Fang, and S. M. Egan. 2007. Transcription activation by the DNA-binding domain of the AraC family protein RhaS in the absence of its effector-binding domain. J. Bacteriol. 1894984-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 81.Zhang, X., T. Reeder, and R. Schleif. 1996. Transcription activation parameters at ara pBAD. J. Mol. Biol. 25814-24. [DOI] [PubMed] [Google Scholar]