Abstract
In the past, studies on the relationships of the bacterial phyla Planctomycetes, Chlamydiae, Lentisphaerae, and Verrucomicrobia using different phylogenetic markers have been controversial. Investigations based on 16S rRNA sequence analyses suggested a relationship of the four phyla, showing the branching order Planctomycetes, Chlamydiae, Verrucomicrobia/Lentisphaerae. Phylogenetic analyses of 23S rRNA genes in this study also support a monophyletic grouping and their branching order—this grouping is significant for understanding cell division, since the major bacterial cell division protein FtsZ is absent from members of two of the phyla Chlamydiae and Planctomycetes. In Verrucomicrobia, knowledge about cell division is mainly restricted to the recent report of ftsZ in the closely related genera Prosthecobacter and Verrucomicrobium. In this study, genes of the conserved division and cell wall (dcw) cluster (ddl, ftsQ, ftsA, and ftsZ) were characterized in all verrucomicrobial subdivisions (1 to 4) with cultivable representatives (1 to 4). Sequence analyses and transcriptional analyses in Verrucomicrobia and genome data analyses in Lentisphaerae suggested that cell division is based on FtsZ in all verrucomicrobial subdivisions and possibly also in the sister phylum Lentisphaerae. Comprehensive sequence analyses of available genome data for representatives of Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes strongly indicate that their last common ancestor possessed a conserved, ancestral type of dcw gene cluster and an FtsZ-based cell division mechanism. This implies that Planctomycetes and Chlamydiae may have shifted independently to a non-FtsZ-based cell division mechanism after their separate branchings from their last common ancestor with Verrucomicrobia.
In the past decade, our view of the bacterial cytoskeleton has basically changed. Bacterial homologues of the three major eukaryotic cytoskeletal families, actin, tubulin, and intermediate filaments, have been identified: FtsA/MreB/ParM, FtsZ/BtubAB, and crescentin (reviewed in reference 9). Another bacterial homologue of tubulin (TubZ) was very recently detected in Bacillus. It was shown to polymerize in dynamic, linear polymers, which were suggested to be important for plasmid maintenance (21). Bacterial cell division is generally based on the tubulin homologue FtsZ, which is the structural protein forming a Z ring at the division site (5). In Escherichia coli, FtsA is the first protein known to be recruited to the Z ring and is thought to be important for stabilization of FtsZ (56, 57). FtsZ is found in almost all bacteria studied so far. However, the analysis of genome-sequencing data suggested that it may be absent in representatives of the three bacterial phyla Verrucomicrobia, Chlamydiae, and Planctomycetes (10, 15, 18, 37, 46).
The report of genuine tubulin genes in the genus Prosthecobacter (18), representatives of Verrucomicrobia, might have suggested a vicarious takeover of the FtsZ function through the bacterial tubulins in the phylum Verrucomicrobia. However, ftsZ genes were recently detected and characterized in three different Prosthecobacter species and in the incomplete genome sequence data for their closest relative, Verrucomicrobium spinosum (36).
Generally, ftsZ is located in a cluster of genes involved in cell division and in the synthesis of peptidoglycan precursors (2, 54). A prominent feature of this division and cell wall (dcw) cluster is that it is conserved in many bacterial genomes over a broad taxonomic range (33, 47, 55). This is remarkable, as genome organization, gene content, and gene order within operons or gene clusters are very dynamic in evolutionary terms (reviewed in reference 11). Further, a strong relationship between the organization/conservation of the dcw cluster and cell morphology has been detected. In contrast to cocci, the majority of rod-shaped and filamentous bacteria have been found to possess a conserved dcw cluster (47). The correlation suggests that cotranslational assembly of some dcw cluster gene products links gene order with cell morphology; this mechanism was referred to as genomic channeling (30, 31). As the last common ancestor of extant bacteria has been suggested to have had a rod shape (20), it was inferred that it also had a large dcw cluster (similar to that of E. coli), which has been maintained in some lineages and has been reduced in others (33). It has been speculated that the ancestral cluster might have arisen from the blending of a primordial mur operon with three other gene subsets, differing in their functional roles (30). Alternatively, but very unlikely, the complex gene structure of the dcw cluster might have been a recent acquisition that has arisen independently by convergent evolution in phylogenetically distant bacterial groups. The possibility that the dcw cluster spread through the bacterial domain by horizontal gene transfer is improbable because the comparative sequence analyses of the individual genes matches the 16S rRNA phylogeny (30). Indeed, genome analysis of deeply branching bacteria also strongly supports the possibility that the last common ancestor of extant bacteria had a large dcw cluster, which was secondarily reduced in different lineages (30).
The dcw cluster comprises 16 genes in E. coli. The first two genes at the 5-prime terminus (mraWZ) code for proteins with unknown functions (6). The mur genes, together with mraY and ddl, are essential and are involved in the synthesis of murein (peptidoglycan) precursors (52). In addition, the dcw cluster comprises six other fts genes that are essential for cell division; their products have been shown to localize at the division site during septation (27). Gene order and content are conserved to different extents in the different regions of the dcw cluster. A variable 5′ region is followed by a conserved block comprising ftsI to murG, followed by another variable region that may contain the genes murC, murB, and ddl; finally, there is another highly conserved block comprising the division genes ftsQAZ (47).
The present knowledge about cell division in the Verrucomicrobia is restricted to the characterization of ftsZ only in the very closely related genera Prosthecobacter and Verrucomicrobium (36, 64). However, the phylum Verrucomicrobia (13, 16) comprises five subdivisions consisting of very heterogeneous free-living and symbiotic species with only very few cultivable representatives.
On the basis of 16S rRNA gene phylogeny, Verrucomicrobia, Chlamydiae, and Planctomycetes form a monophyletic cluster, also comprising Lentisphaera and two additional candidate phyla (Poribacteria and OP3). Despite some studies that disagree (12, 16, 59), the establishment of the new PVC (Planctomycetes-Verrucomicrobia-Chlamydiae) superphylum has been proposed on the basis of 16S rRNA phylogeny (58). We tested the hypothesis of the PVC superphylum using comparative sequence analyses of 23S rRNA genes. We characterized the major bacterial cell division gene ftsZ and other dcw cluster genes in all verrucomicrobial subdivisions with cultivable representatives. Comprehensive sequence analyses and the comparison of the presence of dcw genes and their gene order in representatives of all phyla of the PVC superphylum is interpreted here in relation to the cell division mechanism in their last common ancestor.
MATERIALS AND METHODS
Cultures.
All strains were cultured with shaking in 50 ml liquid media. Cultures of Prosthecobacter dejongeii DSM12251, Prosthecobacter debontii DSM14044, Prosthecobacter vanneervenii DSM12252, and Opitutus terrae DSM11246 were obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Germany). Akkermansia muciniphila CIP107961 was obtained from the Pasteur Institute Collection (France). Chthoniobacter flavus (43) and isolate Ellin514 (44) were obtained from Peter H. Janssen (University of Melbourne, Melbourne, Australia). Prosthecobacter species were grown aerobically at 28°C in DSM medium 628. O. terrae was grown anaerobically at 30°C in DSM medium 298g. A. muciniphila was grown anaerobically at 30°C in brain heart infusion medium (Becton Dickinson and Company). Media for anaerobic cultures were degassed using a vacuum, and the gas atmosphere was exchanged for dinitrogen. C. flavus and strain Ellin514 were grown aerobically at 30°C in medium VL55 (41).
DNA extraction.
DNA from Prosthecobacter was isolated according to the method of Wisotzkey et al. (63). DNA from A. muciniphila, C. flavus, O. terrae, and isolate Ellin514 was extracted using a modified protocol to inhibit high nuclease activity. Liquid cultures were centrifuged, and the pellet was resuspended in TE buffer (10 mM Tris/HCl, 1 mM EDTA disodium salt, pH 8). The lysis of the cells was directly performed in a high sodium dodecyl sulfate concentration without previous lysozyme treatment. Sodium dodecyl sulfate (10%) was added in a 1:1 ratio to the cell resuspension, followed by the addition of proteinase K (0.625 mg/ml) and RNase (0.25 mg/ml). The mixture was incubated for 1 h at 55°C. The subsequent steps were as described by Wisotzkey et al. (63).
Sequencing of 23S rRNA genes.
23S rRNA genes were characterized for P. debontii, P. dejongeii, P. vanneervenii, A. muciniphila, C. flavus, isolate Ellin514, and O. terrae.
Partial rRNA operons were amplified using the primers listed in the supplemental material. The PCR program was as follows: 35 cycles with 30 s of 50°C annealing and 4 min of 72°C extension. Direct sequencing of the 23S rRNA genes was performed by MWG Biotech AG (Germany) using the sequencing primers listed in the supplemental material.
Sequencing of ddl genes.
In our previous study, consensus degenerate hybrid oligonucleotide primers (CODEHOP) (39, 40) were used to detect ftsZ in Prosthecobacter (36). Attempts to detect ftsZ in other Verrucomicrobia using a high number of different combinations of CODEHOP in PCR failed.
Therefore, CODEHOP were designed to target conserved domains of the d-Ala:d-Ala ligase (ddl) gene, which is usually located upstream of the ftsQAZ cluster. A fragment of the ddl gene was initially amplified using the same forward primer for all investigated species, targeting the ddl nucleotide sequences coding for the deduced protein domain GEDG. The reverse primers targeted the ddl nucleotide sequences coding for the deduced protein domains FYDI (P. debontii and P. dejongeii), FYDY (P. vanneervenii), EINT (A. muciniphila), GKEL (C. flavus), and NTIP (strain Ellin514 and O. terrae). Primer sequences are listed in the supplemental material. The PCR program was as follows: 5 cycles with 30 s of 45°C annealing and 1 min of 72°C extension, followed by 30 cycles with 15 s of 55°C annealing and 1 min of 72°C extension. PCR products were analyzed on 2% agarose gels. Selected fragments were cut from the gel, and DNA was extracted using the Perfectprep kit (Eppendorf, Germany). Fragments were cloned using a commercially available TOPO TA cloning kit (Invitrogen), and inserts were sequenced by MWG Biotech AG (Germany) using vector-specific primers.
Sequencing of the genomic environment of ddl and ftsZ.
In P. dejongeii and P. debontii, specific primers targeting ddl and ftsZ were used in PCR to specifically amplify and subsequently sequence the intervening region comprising ftsQA. For the remaining investigated Verrucomicrobia, two-step gene walking, as described by Pilhofer et al. (35), was applied to characterize the genomic environment of the ddl gene. Attempts to walk downstream of the O. terrae ddl gene were not performed, as there are two ongoing genome projects on Opitutaceae isolates. The sequencing results were verified, and chimera formation during gene walking was excluded using both specific primers and genomic DNA as templates in PCR and Southern hybridization experiments (data not shown). The procedure was described previously (35).
Detection of dcw cluster genes in genome project data.
The Integrated Microbial Genomes System (28) and BLASTP (1) were used to screen for the presence of dcw cluster genes in available genome project data.
At present, genome sequence data (unpublished genomes) for five isolates and strains belonging to Verrucomicrobia or Lentisphaerae (the closest relatives of Verrucomicrobia) are publicly available. The strains are V. spinosum (Verrucomicrobia, subdivision 1, Verrucomicrobiae), isolate DG1235 (Verrucomicrobia, subdivision 4, Opitutaceae), isolate TAV2 (Verrucomicrobia, subdivision 4, Opitutaceae), Lentisphaera araneosa (Lentisphaerae), and Victivallis vadensis (Lentisphaerae). The sequence data for isolate TAV2 and Victivallis vadensis were produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/). Preliminary sequence data for isolate DG1235 and L. araneosa were obtained from the J. Craig Venter Institute or from the Gordon and Betty Moore Foundation Marine Microbial Genome Sequencing Project website (https://research.venterinstitute.org/moore/).
For Chlamydiae, the published genomes of Chlamydia pneumoniae, Chlamydia trachomatis, Chlamydophila caviae, Chlamydophila felis, Chlamydophila abortus, and “Candidatus Protochlamydia amoebophila” were analyzed (3, 15, 19, 37, 38, 51). For Planctomycetes, the genomes of Rhodopirellula baltica (10), “Candidatus Kuenenia stuttgartiensis” (46), Planctomyces maris (unpublished), and Blastopirellula marina (unpublished) were analyzed. Preliminary sequence data for Planctomyces maris and Blastopirellula marina were obtained from the J. Craig Venter Institute website (http://www.jcvi.org/).
RNA isolation and transcription analyses.
Total RNA was isolated from Prosthecobacter cultures (20 ml; mid-log phase) by using Trizol reagent (Invitrogen). The protocol of the manufacturer was modified. After cells were pelleted by centrifugation, the cells were lysed by adding 1 ml Trizol reagent. The cells were resuspended by vortexing them for 30 s and then incubated for 5 min at 20°C and for 1 h at −80°C. The subsequent steps were as described in the instructions of the manufacturer.
DNase (RQ1 RNase-Free DNase; Promega) treatment of total RNA was performed according to the instructions of the manufacturer. Subsequently, reverse transcription was carried out with the Revert Aid First Strand cDNA kit (Fermentas, Germany) according to the instructions of the manufacturer and by using random hexamer primers. One microliter of the obtained cDNA was used for PCR (25-μl final volume), together with specific primers (see the supplemental material) for the genes ddl, ftsQ, ftsA, and ftsZ.
Sequence analyses.
Open reading frames (ORFs) were detected using the ORF-Finder at http://www.ncbi.nlm.nih.gov (62). Similarities to known protein sequences and to conserved protein domains were searched using protein-protein BLAST (1). The program MOTIF Search (http://motif.genome.jp/) was used in order to detect conserved domains in Ddl and FtsQAZ protein sequences.
Phylogenetic sequence analyses.
Comparative sequence analyses were performed using the ARB program package (24). The 23S rRNA ARB database comprised the new 23S rRNA sequences obtained in this study and sequences of the major bacterial groups. For tree calculation, 96 (860 for RAxML) sequences were selected representing all available sequences of Verrucomicrobia, Lentisphaerae, Chlamydiae, Planctomycetes, and selected species of the major bacterial groups, some Archaea, and some 28S rRNA sequences of Eukarya. Minimum-similarity filters were calculated, which retained only positions conserved in at least 0%, 20%, or 50% of the selected sequences. The phylogenetic analyses were performed using each filter in combination with each treeing program. The treeing methods used were distance matrix methods (Phylip NEIGHBOR and FITCH), maximum parsimony (Phylip DNAPARS), and maximum likelihood (RAxML, AxML, and PHYML DNA). All programs are implemented in the ARB program package (24). Maximum-parsimony and maximum-likelihood trees were bootstrap resampled (1,000 bootstraps). As a nucleotide substitution model, the Kimura-2-Parameter was used for distance matrix methods and PHYML; the HKY85 model was used for RAxML.
The FtsZ_ClustalW ARB database published by Pilhofer et al. (36) was extended with the new sequences obtained in this study. A Ddl ARB database was established importing the Ddl protein sequences obtained in this study and Ddl proteins of other major bacterial groups. The initial alignment was produced by using ClustalW (50) of the ARB program package (24) and was refined manually. Minimum-similarity filters were calculated in each database, which retained only positions conserved in at least 0% or 30% of the selected sequences, respectively. Phylogenetic analyses were performed using distance matrix methods (the programs ARB neighbor joining and Phylip UPGMA, FITCH, and KITSCH), maximum parsimony (Phylip PROTPARS), and maximum likelihood (Phylip PROML and TREE-PUZZLE [45]). All programs are implemented in the ARB program package (24). For distance matrix methods and the maximum-likelihood method, the model of substitution used was the Dayhoff PAM matrix (8); for the TREE-PUZZLE method, the Muller-Vingron model of substitution (32) was used. Maximum-parsimony trees were bootstrap resampled (10,000 bootstraps); the number of puzzling steps for TREE-PUZZLE trees was 100,000.
Nucleotide sequence accession numbers.
Nucleotide sequences were submitted to the EMBL nucleotide database with accession numbers AJ96882 to AJ96884 and AM905423 to AM905426. All characterized nucleotide sequences of the dcw clusters were submitted to the EMBL nucleotide database with accession numbers AM905291 to AM905296. Entries AJ888907 and AJ888908 were updated.
RESULTS
Phylogenetic sequence analyses of 23S rRNA genes.
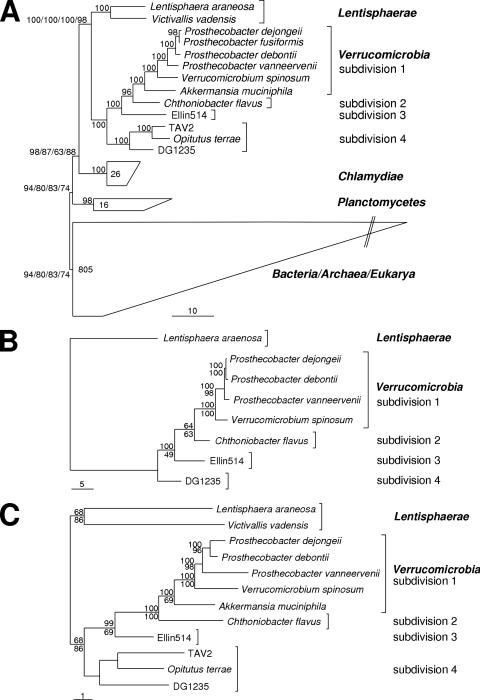
23S rRNA genes were characterized for P. dejongeii, P. debontii, P. vanneervenii, A. muciniphila, C. flavus, Ellin514, and O. terrae. For the remaining investigated Verrucomicrobia and Lentisphaerae, 23S rRNA gene sequences were retrieved from the genome project data. The data set used for calculation of phylogenetic trees included all available 23S rRNA sequences of members of the Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes and selected representatives of the major bacterial groups, some Archaea, and some Eukarya. Trees were calculated using 0%, 20%, and 50% minimum-similarity filters in combination with the different treeing methods (see Materials and Methods). (For one representative maximum-likelihood tree, see Fig. 4A; a further selection of trees is available as supplemental material.) All 18 calculated trees showed a monophyly of all included members of the PVC superphylum. The tree topology within the PVC superphylum was also stable, showing the monophyletic groups always branching in the order Planctomycetes, Chlamydiae, Lentisphaerae, and Verrucomicrobia. The verrucomicrobial subdivisions (1 to 4) also showed a monophyletic and stable treeing pattern.
FIG. 4.
Comparative sequence analyses using 23S rRNA (A), FtsZ (B), and Ddl (C). All calculated 23S rRNA trees showed a stable monophyletic group of Planctomycetes, Chlamydiae, Lentisphaerae, and Verrucomicrobia with a stable branching order within this PVC superphylum. Verrucomicrobial subdivisions were recovered monophyletically and showed a branching order matching the 16S rRNA trees. Other 23S rRNA trees are available in the supplemental material. Verrucomicrobial FtsZ and Ddl trees show the same tree topologies as 16S and 23S rRNA trees. (A) Phylogenetic 23S rRNA tree produced using the maximum-likelihood method (RAxML) and a minimum-similarity filter (50%). Selected representatives of major bacterial groups, Archaea, and Eukarya were used as outgroups. Note that the tree is unrooted. The branching pattern within the group “Bacteria/Archaea/Eukarya” is shown in the supplemental material. The numbers in closed groups denote the numbers of sequences included. The numbers at the nodes represent bootstrap values (1,000 bootstraps) (percent). Four bootstrap values are shown for the branchings of the phyla (maximum likelihood, 0% minimum-similarity filter; maximum likelihood, 50% minimum-similarity filter; maximum parsimony, 0% minimum-similarity filter; maximum parsimony, 50% minimum-similarity filter); one bootstrap value is shown for the remaining nodes (maximum parsimony, 50% minimum-similarity filter). The bar represents the estimated evolutionary distance (percent). (B) Phylogenetic FtsZ (core domain) tree produced using the maximum-likelihood method and a minimum-similarity filter (30%). The numbers represent bootstraps/confidence values of a maximum-parsimony tree (lower numbers, 10,000 bootstraps) and a TREEPUZZLE tree (upper numbers, 100,000 puzzling steps) (percent). The bar represents the estimated evolutionary distance (percent). (C) Phylogenetic Ddl tree produced using the maximum-likelihood method and a minimum-similarity filter (30%). The numbers represent bootstraps/confidence values of a maximum-parsimony tree (lower numbers, 10,000 bootstraps) and a TREEPUZZLE tree (upper numbers, 100,000 puzzling steps) (percent). The bar represents the estimated evolutionary distance (percent).
Overall, the 23S rRNA trees were in absolute agreement with many published 16S rRNA phylogenies (7, 16, 43, 58) in regard to the monophyly of the PVC superphylum, the branching order of the different phyla within the PVC superphylum, and the tree topology within Verrucomicrobia.
Detection and genomic organization of ddl and ftsQAZ in Verrucomicrobia and Lentisphaerae.
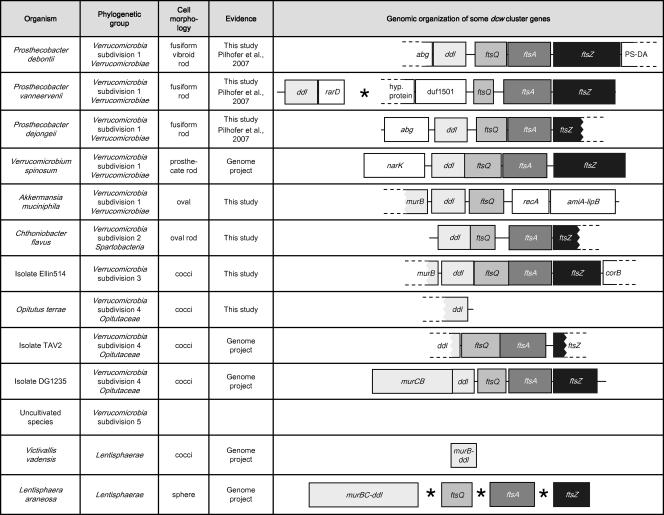
Sequences with similarities to ddl and ftsZ could be detected in all verrucomicrobial subdivisions with cultivable representatives.
With the exception of P. vanneervenii and A. muciniphila, the intervening region of ddl-ftsZ (generally ftsQA) was shown to be conserved (Fig. 1). Downstream of the P. vanneervenii ddl gene, an ORF with similarities to rarD genes (coding for predicted permeases) (26) could be detected. Upstream of the complete ftsQ sequence, an ORF with similarities to the conserved protein domain duf1501, a protein family with unknown function, was identified (Fig. 1). In A. muciniphila, ftsQ was detected downstream of the ddl gene, followed by a gene with similarities to recA (a bacterial recombination protein). Downstream of the recA homolog, a single ORF with similarities to both amiA (N-acetylmuramoyl-l-alanine amidase; N terminal) and lipB (lipoyltransferase; C terminal) was detected (Fig. 1).
FIG. 1.
Genomic organization of the 3′-terminal part of the dcw cluster in Verrucomicrobia and Lentisphaerae. The genes shaded in gray or black are generally organized within the dcw cluster. The genes in white are not functionally related to and normally not organized within the dcw cluster. The asterisk indicates that contigs are isolated on the chromosome. The dashed lines indicate that the sequence of the ORF is partial. With two exceptions, Verrucomicrobia show the conserved gene order ddl-ftsQAZ, whereas in Lentisphaerae, the individual genes are isolated on the chromosome.
ddl, ftsQ, ftsA, and a sequence with some similarities to ftsZ were also detected in L. araneosa (Lentisphaerae, closest relatives of Verrucomicrobia), but the single genes were organized isolated at different locations on the genome (Fig. 1). In the incomplete V. vadensis (Lentisphaerae) genome sequence data, only the ddl gene could be identified (Fig. 1).
Genomic organization of dcw cluster genes in the phyla Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes.
The upstream region of ddl in Verrucomicrobia/Lentisphaerae showed either murB (A. muciniphila and Ellin514), a murCB fusion (DG1235), or the ddl gene fused with murB or murBC (V. vadensis and L. araneosa, respectively) (Fig. 1). In some other cases the upstream region was not conserved and apparently not related to the dcw cluster (P. debontii, P. dejongeii, and V. spinosum) (Fig. 1).
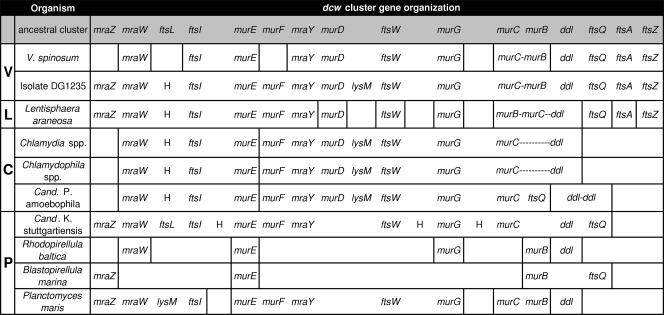
The only verrucomicrobial organism for which the available sequence data could prove the conservation of the gene order of the complete dcw cluster was isolate DG1235 (Fig. 2). Notably, the gene content and order were highly similar to the proposed ancient dcw cluster type (Fig. 2) (48). The only differences from the ancient type were (i) the replacement of ftsL through a hypothetical protein, (ii) the insertion of lysM between murD and ftsW, and (iii) the murC-murB fusion (Fig. 2). The first feature is shared with L. araneosa and all Chlamydiae; the second feature is shared only with Chlamydiae.
FIG. 2.
Genomic organization of dcw genes in Verrucomicrobia, Lentisphaerae, Chlamydiae, and Planctomycetes. Comparison of dcw gene content and gene order conservation in the PVC superphylum. The hypothetical ancestral cluster (47) is shown in the first line. The first column designates phylogenetic groups: V, Verrucomicrobia; L, Lentisphaerae; C, Chlamydiae; P, Planctomycetes. The vertical lines indicate isolated organization on the chromosome. Hyphens denote gene fusions. H stands for hypothetical protein. DG1235 shows a good conservation of the dcw cluster with some peculiarities compared to the ancestral type. L. araneosa presents most of the dcw genes, whereas the gene order is lost. Chlamydiae dcw clusters are disrupted in two parts but are remarkably similar to DG1235, except that they contain no fts(Q)AZ genes. Planctomycetes show high variability in the conservation of gene content and gene order.
Ongoing and finished genome projects of the phyla Lentisphaerae, Chlamydiae, and Planctomycetes were also investigated for the presence of dcw cluster genes. With the exception of ftsL, L. araneosa was found to also possess all genes of the proposed ancient dcw cluster; however, the genes generally located in the 3′-terminal region of the cluster (ftsQ, ftsA, and ftsZ-like) were dispersed in the genome (Fig. 2).
In Chlamydiae representatives, ftsZ, ftsA, ftsQ (with the exception of “Ca. Protochlamydia”), ftsL, and mraZ are absent. The remaining dcw genes are arranged in two isolated clusters, which are remarkably more similar to DG1235 than to the ancestral cluster (ftsL replaced by a gene coding for a hypothetical protein; lysM insertion). Chlamydia and Chlamydophila show a fused murC-ddl gene, whereas in “Ca. Protochlamydia,” murC is followed by ftsQ and the ddl gene is isolated (Fig. 2).
In representatives of Planctomycetes, the dcw gene content and order conservation vary widely between the different representatives. Most of the genes of the ancient dcw cluster are present in “Candidatus Kuenenia stuttgartiensis.” They are organized in a cluster that is very similar to that predicted for the ancestral type; ftsA, ftsZ, murD, and murB are absent, and three ORFs coding for hypothetical proteins are inserted. In contrast, R. baltica and B. marina genome data present only some isolated mur genes and none of the fts genes (ftsQ in B. marina). P. maris possesses most of the dcw genes, but they are clustered at different locations in the genome (Fig. 2).
Transcription analyses.
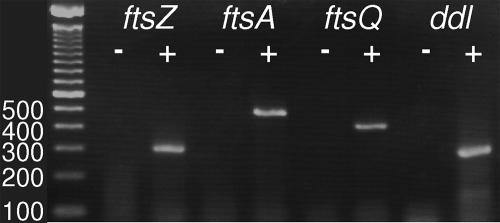
The transcription of ddl, ftsQ, ftsA, and ftsZ was verified in P. dejongeii and P. vanneervenii using reverse transcription of mRNA (Fig. 3). Total RNA was isolated from Prosthecobacter cultures and was subsequently treated using DNase. Total RNA was reverse transcribed using random hexamers. The cDNA was subsequently used as a template in PCR with gene-specific primers. Control reactions for each PCR were one reaction without template and one control sample processed without reverse transcriptase (Fig. 3). Specific amplification from cDNA was detected for all investigated genes in both species.
FIG. 3.
Transcriptional analyses of dcw cluster genes in Prosthecobacter. Five-microliter aliquots of PCR products were loaded on a 2% agarose gel. The template for PCR was the cDNA sample (+) or a control sample processed without reverse transcriptase (−). A 100-bp ladder (Invitrogen) was loaded on the left side; the numbers refer to fragment lengths in base pairs. The expected sizes of the fragments were 296 bp (ftsZ), 435 bp (ftsA), 354 bp (ftsQ), and 283 bp (ddl). Transcription was detected for all investigated genes in P. dejongeii (shown) and P. vanneervenii.
Search for conserved protein domains and residues.
In order to verify the sequence similarities detected using BLASTP, Ddl and FtsQAZ protein sequences of Verrucomicrobia/Lentisphaerae were searched for conserved domains of the Pfam library (4). The matching scores of the query sequences to the respective Pfam domains are listed as E values in Table 1. All detected Ddl sequences showed matches to both d-Ala:d-Ala ligase Pfam domains with comparable high scores, except for two partial sequences (O. terrae and TAV2). For all available investigated FtsQA sequences, there were matches to the POTRA domain FtsQ type (42) or to the cell division protein FtsA domain, respectively. Due to the high sequence divergence of these proteins, the scores for the matches were generally lower than Ddl matches (Table 1). All complete verrucomicrobial FtsZ sequences showed a match to the tubulin/FtsZ family GTPase domain, with comparable high scores. The sequence of L. araneosa (Lentisphaerae) with similarities to FtsZ did not match the tubulin/FtsZ family GTPase Pfam domain, nor did it match the FtsZ PROSITE pattern, the BLOCKS database, or the PRINTS motif (the search was performed using MOTIF search [http://motif.genome.jp/]). However, the sequence was initially detected using BLASTP and showed similarities to the conserved domain FtsZ (cd02191) (26) and to FtsZ from P. debontii with E values of 5e−06 and 6e−04, respectively.
TABLE 1.
E valuesa of dcw genes for Pfam domains
| Isolate | E value
|
||||
|---|---|---|---|---|---|
| Ddl
|
FtsQ (POTRA domain; FtsQ type) | FtsA (cell division protein FtsA) | FtsZ (tubulin/FtsZ family; GTPase) | ||
| d-Ala:d-Ala ligase C terminus | d-Ala:d-Ala ligase N terminus | ||||
| P. dejongeii | 9.8e−53 | 2.1e−15 | 0.01 | 1.3e−38 | 5.9e−49b |
| P. debontii | 2.9e−59 | 1.5e−16 | 0.0042 | 7.8e−40 | 3.3e−50 |
| P. vanneervenii | 2.3e−64 | 2.0e−14 | 4.7e−11 | 8.9e−42 | 5.8e−54 |
| V. spinosum | 2.0e−69 | 2.1e−11 | 1.5e−07 | 1.3e−42 | 5.4e−48 |
| A. muciniphila | 3.5e−56 | 2.8e−18 | 7.9e−06 | ||
| C. flavus | 9.7e−54 | 3.4e−13 | 5.4e−10 | 2.4e−50 | 5.5e−60b |
| Ellin514 | 3.5e−57 | 3.7e−19 | 1.8e−09 | 2.0e−46 | 7.1e−67 |
| O. terrae | 1.2e−48b | −b,c | |||
| TAV2 | −b,c | −b,c | 5.4e−12 | 1.2e−19 | 0.27b |
| DG1235 | 2.6e−45 | 1.3e−13 | 4.5e−12 | 1.7e−24 | 1.2e−53 |
| V. vadensis | 5.8e−53 | 9.6e−21 | |||
| L. araneosa | 3.4e−47 | 1.3e−26 | 3.4e−15 | 7.0e−05 | −c |
| E. coli | 3.4e−119 | 2.9e−47 | 8.7e−37 | 6.7e−79 | 7.8e−91 |
Based on scoring matches to the motifs.
Partial sequence.
No significant hit.
FtsZ sequences were additionally searched for the tubulin signature motif (S/A/G)GGTG(S/A/T)G (PROSITE motif PS00227), which is present and perfectly conserved in all investigated verrucomicrobial FtsZ proteins. In contrast, the putative L. araneosa FtsZ homolog did not show the tubulin signature motif. A similar observation was made during the analysis of amino acid positions that contact GDP (23, 34). The analysis performed by Pilhofer et al. (36) was extended using the newly obtained FtsZ sequences (data not shown). All important positions were conserved among Verrucomicrobia, even if the new FtsZ sequences of subdivisions 2 to 4 were added. The L. araneosa FtsZ-like protein showed no conservation in the majority of these amino acid residues. BLASTP analysis of ddl ORFs revealed that Verrucomicrobia show the common structure comprising one Ddl domain. Whereas, similar to ddl genes of Verrucomicrobia, ddl genes of Planctomycetes comprise only one Ddl domain, Ddls of Lentisphaerae and Chlamydiae present different types of fused genes (Fig. 2). ddl genes were found to be fused with murBC (L. araneosa), murB (V. vadensis), and murC (Chlamydia and Chlamydophila) or with another ddl (“Ca. Protochlamydia”).
In BLASTP analyses, the first hits detected for Verrucomicrobia, Planctomycetes, Chlamydia, and Chlamydophila Ddl sequences were other bacterial Ddls (data not shown). Notably, the “Candidatus Protochlamydia amoebophila” sequence comprising two Ddl domains showed the best matches (E value, 3e−161) to a nuclear-encoded gene in Physcomitrella patens (Viridiplantae), a moss for which ddl and related genes seem to be important for plastid division (25).
Phylogenetic analyses using Ddl and FtsZ sequences.
The newly obtained FtsZ and Ddl sequences were used for phylogenetic-tree reconstructions. The FtsZ_ClustalW ARB database published by Pilhofer et al. (36) was extended with the new sequences obtained in this study. FtsZ sequences of Verrucomicrobia and Lentisphaerae (excluding partial TAV2 FtsZ) were used to produce two different minimum-similarity filters. Phylogenetic trees were calculated using both filters in combination with distance matrix methods, maximum parsimony, and maximum likelihood. Attempts to calculate global trees could recover a relationship of the L. araneosa FtsZ-like protein and verrucomicrobial FtsZs in most cases, whereas the relationships between the other major bacterial groups were unstable (data not shown; compare reference 36). Therefore, subsequent phylogenetic analyses were restricted to Lentisphaerae and Verrucomicrobia. All calculated trees showed stable tree topologies, with stable monophyletic groups representing the different subdivisions. In all trees, the branching patterns of the FtsZ sequences of the different verrucomicrobial subdivisions (1, 2, 3, and 4) were consistent with the pattern obtained in rRNA phylogeny (Fig. 4) (7, 16, 43, 58). One representative maximum-likelihood tree is shown in Fig. 4B.
A Ddl ARB database was established by importing the Ddl protein sequences obtained in this study and Ddl sequences of other bacterial groups from the EMBL nucleotide database. Initial attempts to calculate global trees using different methods and filters showed that in most cases the major bacterial groups could be recovered, whereas the relationships between the major bacterial groups were unstable (data not shown), as is also the case for FtsZ phylogeny (36). Therefore, subsequent phylogenetic analyses were restricted to Lentisphaerae and Verrucomicrobia. Notably, all global Ddl trees showed a stable relationship of “Candidatus Protochlamydia amoebophila” Ddl with P. patens (Viridiplantae) Ddl (nuclear encoded). The “Ca. Protochlamydia” Ddl was duplicated in the database, so each of the Ddl domains could be aligned once.
Ddl sequences of Verrucomicrobia and Lentisphaerae (excluding partial TAV2 Ddl) were used to calculate two minimum-similarity filters. Phylogenetic trees were reconstructed using both filters in combination with distance matrix methods, maximum parsimony, and maximum likelihood. As the isolate TAV2 Ddl sequence is partial and therefore very short, it was added separately to each tree using ARB Interactive Parsimony and a special filter considering the length of TAV2 Ddl. One representative maximum-likelihood tree is shown in Fig. 4C. All calculated trees showed stable tree topology, with stable monophyletic groups representing the two phyla Verrucomicrobia and Lentisphaerae. Within the Verrucomicrobia, each subdivision (1, 2, 3, and 4) was recovered monophyletically; however, the bootstrap support for the subdivision 4 cluster was not high. The branching pattern is consistent with the pattern obtained using comparative sequence analyses of 16S rRNA genes (7, 16, 43, 58), 23S rRNA genes (Fig. 4A), and FtsZ proteins (Fig. 4B).
DISCUSSION
Phylogenetic relationships between Planctomycetes, Chlamydiae, Lentisphaerae and Verrucomicrobia.
A stable monophyly of Planctomycetes, Chlamydiae, Lentisphaerae and Verrucomicrobia could be detected using 23S rRNA phylogeny (Fig. 4A). Also, the bootstrap support was high for this clade (14). In the past, there were also other studies, addressing the phylogenetic relationships between Planctomycetes, Chlamydiae, and Verrucomicrobia based on 16S rRNA (7, 13, 22, 58, 61), 23S rRNA (59), or protein sequences (12, 17, 46, 49, 60), in which the formation of a monophyletic cluster of all three lineages was controversial. The study by Ward et al. (59), also using 23S rRNA as a phylogenetic marker, could not detect a significant relationship of the three phyla. However, the data set was rather limited: Planctomycetes were represented by 13 sequences (16 representatives in this study), Chlamydiae by one sequence (26 representatives in this study), and Verrucomicrobia by three sequences (11 representatives in this study), and Lentisphaerae were completely lacking (2 representatives in this study). The much more comprehensive phylogenetic analyses in this study showed very good agreement between trees derived from 23S and 16S rRNAs (Fig. 4). Therefore, these analyses support the suggestion of a PVC superphylum based on 16S rRNA phylogeny in which the sister phyla Lentisphaerae and Verrucomicrobia are more closely related to Chlamydiae than to Planctomycetes (58).
Origin of the “Candidatus Protochlamydia amoebophila” ddl-ddl gene.
An intriguing feature derives from the analysis of “Candidatus Protochlamydia” Ddl-Ddl. This gene shows unexpectedly high similarity to plant Ddl-Ddl, involved in chloroplast division (25), and apparently low similarity to bacterial Ddl. The plant ddl genes have been characterized up to now in Arabidopsis, in Oryza, and in the moss Physcomitrella. For the first two organisms, genomic data are available, and they show the presence of many introns interrupting the coding region of the ddl genes. The observed high similarity between plants and “Ca. Protochlamydia” ddl suggests that either plants acquired ddl from a “Ca. Protochlamydia” ancestor and later inserted introns in the coding region or “Ca. Protochlamydia” acquired ddl from plants, most likely via reverse-transcription-mediated mechanisms (due to the “necessity” to remove introns). The latter case would imply that some bacterial genes of the dcw cluster might be lost and reacquired through horizontal gene transfer.
FtsZ-based cell division mechanism in Verrucomicrobia and Lentisphaerae.
In contrast to previous assumptions, ftsQAZ and ddl genes could be detected in representatives of all verrucomicrobial subdivisions. The failure to detect ftsZ in A. muciniphila and O. terrae by PCR is not significant, due to the sequence divergence of ftsZ (see Materials and Methods); we predict that these organisms also possess ftsZ. Phylogenetic sequence analyses using the obtained FtsZ and Ddl sequences are in full agreement with the rRNA trees (Fig. 4) (7, 16, 43, 58). This consistency suggests the absence of horizontal gene transfer influence on FtsZ and Ddl evolution in Verrucomicrobia and strongly indicates that the genes are conserved due to functional constraints. This also applies to the isolated genes of the nonconserved dcw clusters (in terms of gene order) of P. vanneervenii and A. muciniphila (Fig. 1). Comprehensive analyses of the FtsZ sequences revealed that all verrucomicrobial FtsZ proteins matched the criteria of functional FtsZ as defined by Vaughan et al. (53). The sequences showed a match to the conserved Pfam domain tubulin/FtsZ family GTPase, the tubulin signature motif could always be detected without mismatches, and the analysis of amino acid positions that contact GDP (23, 34) also showed good conservation. The constant presence of ftsQAZ in Verrucomicrobia, the phylogenetic conservation of FtsZ, the fulfilment of all criteria of functional FtsZ within the gene sequence, and the proof of transcription of ftsQAZ in actively growing cultures provide a strong indication of functionality and suggest an FtsZ-based cell division mechanism in all members of the phylum Verrucomicrobia.
Concerning the phylum Lentisphaerae, represented by the cultivated species V. vadensis and L. araneosa, the situation is less clear. Considering that the genome sequence of V. vadensis is still unfinished (324 contigs), the present inability to detect ftsQAZ-like sequences in the genome data is not significant. Indeed, its closest relative, L. araneosa, possesses ftsQ, ftsA, and an ftsZ-like gene, although all of them are spread out among different positions on the chromosome. Concerning L. araneosa FtsZ, similarities to Prosthecobacter FtsZ and to the conserved domain FtsZ could be clearly detected, whereas the criteria of functional FtsZ (53) are not fulfilled at all. L. araneosa FtsQA have matches to the respective Pfam motifs with high E values (Table 1), which could be an indication of functionality. Up to now, no bacteria that had a non-FtsZ-based cell division mechanism and at the same time possessed ftsA (encoding the first protein recruited to the Z ring and directly interacting with FtsZ) could be identified. In summary, the present data may suggest that Lentisphaerae also perform FtsZ-based cell division like Verrucomicrobia and the majority of other bacteria. However, the failure to meet the criteria of functionality could indicate a modified mechanism or even that the gene is (at least in cell division) nonfunctional. Future studies of the L. araneosa FtsZ-like protein could verify whether the fundamental criteria of functional FtsZ (53) have to be revised.
Conservation of the gene content and gene order of the dcw cluster in Verrucomicrobia and Lentisphaerae.
The analyses of the available sequence data demonstrated that in at least one verrucomicrobial organism (DG1235) the dcw cluster is nearly perfectly conserved in comparison to the ancient type of cluster, which comprises 16 genes (Fig. 2) (48). In at least five other cases (all investigated subdivision 1 organisms), gene order in the cluster showed variations (Fig. 1). With one exception (ftsL), Lentisphaera maintained the gene content, whereas the gene order was conserved only in the 5′-terminal part of the dcw cluster (Fig. 2).
A comparison of the dcw clusters from several bacterial genomes showed that there is a relationship between the organization of the gene cluster and the shape of the cells, so that the cluster tends to be conserved in rod-shaped and filamentous bacteria and tends to be disrupted in bacteria with other shapes (48). This observation is supported in this study, as none of the Verrucomicrobia/Lentisphaerae with dispersed dcw cluster genes formed classical cylindrical rods with hemispherically capped ends. The types of morphology in Verrucomicrobia/Lentisphaerae range from spheres (L. araneosa), cocci (V. vadensis, DG1235, TAV2, O. terrae, and Ellin514), oval rods (C. flavus), oval shapes (A. muciniphila), and prosthecate rods (V. spinosum) to fusiform rods (Prosthecobacter) (Fig. 1). It is notable that there are two different types of dcw cluster organizations (concerning the generally highly conserved 3′ region) within the genus Prosthecobacter that have very similar morphologies (P. vanneervenii-P. dejongeii and P. debontii) (Fig. 1). Mingorance et al. (30, 31) proposed a model, called genomic channeling, in which the selective pressure to maintain the dcw cluster arises from the need to efficiently coordinate the process of elongation and septation in rod-shaped bacteria. Although Prosthecobacter and Verrucomicrobium have rod-like morphologies, they also possess one or many prosthecae. It can be suspected that the presence of these prosthecae is likely to force a growth mechanism rather divergent from the one found in classical rods. Thus, it is hard to predict if and how peptidoglycan elongation and septation machineries compete. Therefore, it seems likely that genomic channeling of the dcw cluster does not play a major role in these organisms, and it is very probable that other mechanisms for the determination of cell shape exist in Prosthecobacter.
Conservation of gene content and gene order of the dcw cluster in Planctomycetes and Chlamydiae compared to Verrucomicrobia/Lentisphaerae.
The high similarity of dcw clusters within Chlamydiae could be derived from adaptation to their similar intracellular lifestyles and to the consequent functional constraints. Although up to now the existence of peptidoglycan in Chlamydiae has not been proven, the genes coding for a nearly complete pathway for peptidoglycan synthesis were suggested to be functional (reviewed in reference 29). In contrast to Chlamydiae, Planctomycetes show a high degree of variability in the dcw cluster, which could be related to the very different lifestyles of its representatives.
The hypothesis that the last common ancestor of extant bacteria harbored a conserved dcw cluster and that some lineages lost this gene order during evolution (33) is strongly supported by the observations obtained in this study. In comparison to the ancestral dcw cluster type, DG1235 shows only one gene replacement, one gene insertion, and one gene fusion; Chlamydiae show the gene order conservation of many dcw genes; and 12 out of 16 genes are conserved in the Planctomycetes representative “Candidatus Kuenenia stuttgartiensis” (Fig. 2). These results are also a strong indication that the last common ancestor of the PVC superphylum possessed a conserved dcw cluster. Combined with the phylogenetic analyses, these data clearly support the view that many representatives of Planctomycetes, Chlamydiae, and Lentisphaerae independently lost some of the dcw genes or the gene order within the dcw cluster. An exception to this might be the replacement of ftsL by a hypothetical protein in DG1235 (Verrucomicrobia), L. araneosa (Lentisphaerae), and all Chlamydiae; this replacement might represent a characteristic of the last common ancestor of Verrucomicrobia-Lentisphaerae-Chlamydiae. In the same way, the insertion of lysM between murD and ftsW might represent a synapomorphic feature of Verrucomicrobia and Chlamydiae (the absence of lysM in V. spinosum in the expected position could be interpreted as a secondary loss).
In the scenario postulated here, after the separate branchings of Chlamydiae and Planctomycetes from their last common ancestor with Verrucomicrobia, their cell division mechanisms most probably shifted independently from an FtsZ-based mechanism to a non-FtsZ-based mechanism. The major cell division gene ftsZ was completely lost or diverged so far from the original ftsZ that a relationship is no longer recognizable. FtsZ could have been replaced by two different division mechanisms (concerning the structural protein of the division ring) in the different phyla and therefore does not necessarily have to be homologous. Another possibility is that there was an independent shift to the same division mechanism in Chlamydiae and Planctomycetes. If this was the case, it seems probable that the mechanism could have already been present in the last common ancestor of Planctomycetes/Chlamydiae/Lentisphaerae/Verrucomicrobia. To the best of our knowledge, two Ureaplasma species are the only examples in bacteria besides Chlamydiae and Planctomycetes that were identified to lack ftsZ. Thus, a special predisposition of the last common ancestor of the PVC superphylum to evolve an FtsZ-independent type of cell division can be hypothesized.
The single presently identifiable L. araneosa ftsZ-like gene (in the unfinished genome sequence) shows the highest similarities to verrucomicrobial FtsZs. At the same time, the sequence divergence in comparison to other bacterial FtsZ proteins is higher than that of verrucomicrobial FtsZ. Thus, from an evolutionary perspective, L. araneosa could be seen as an organism on the way to developing an FtsZ-independent cell division mechanism or one that has already developed such a mechanism.
Supplementary Material
Acknowledgments
We thank Peter H. Janssen for providing us the bacterial strains Chthoniobacter flavus and isolate Ellin514. Tom Schmidt is acknowledged for the information about the morphology of isolate TAV2. Vladimir Zverlov is acknowledged for assistance with anaerobic cultures. The U.S. Department of Energy Joint Genome Institute, the J. Craig Venter Institute, and the Gordon and Betty Moore Foundation Marine Microbial Genome Sequencing Project are acknowledged for providing preliminary sequence data on their home pages.
This work was supported by the Bayerische Forschungsstiftung (fellowships to M.P. and K.R.), the German Research Organization (grant Schl 120/15), and the Italian Research Ministry (PRIN protocol 2006053200_002).
Footnotes
Published ahead of print on 29 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayala, J. A., T. Garrido, M. A. Pedro, and M. Vicente. 1994. Molecular biology of bacterial septation. In J. M. Ghuysen and R. Habenbeck (ed.), Bacterial cell wall. Elsevier Science B.V., Amsterdam, The Netherlands.
- 3.Azuma, Y., H. Hirakawa, A. Yamashita, Y. Cai, M. A. Rahman, H. Suzuki, S. Mitaku, H. Toh, S. Goto, T. Murakami, K. Sugi, H. Hayashi, H. Fukushi, M. Hattori, S. Kuhara, and M. Shirai. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 1315-23. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, E., and J. Lutkenhaus. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354161-164. [DOI] [PubMed] [Google Scholar]
- 6.Carrion, M., M. J. Gomez, R. Merchante-Schubert, S. Dongarra, and J. A. Ayala. 1999. mraW, an essential gene at the dcw cluster of Escherichia coli codes for a cytoplasmic protein with methyltransferase activity. Biochimie 81879-888. [DOI] [PubMed] [Google Scholar]
- 7.Cho, J. C., K. L. Vergin, R. M. Morris, and S. J. Giovannoni. 2004. Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae. Environ. Microbiol. 6611-621. [DOI] [PubMed] [Google Scholar]
- 8.Dayhoff, M. O., R. M. Schwartz, and B. C. Orcutt. 1978. Atlas of protein sequence and structure. National Biomededical Research Foundation, Silver Spring, MD.
- 9.Gitai, Z. 2007. Diversification and specialization of the bacterial cytoskeleton. Curr. Opin. Cell Biol. 195-12. [DOI] [PubMed] [Google Scholar]
- 10.Glockner, F. O., M. Kube, M. Bauer, H. Teeling, T. Lombardot, W. Ludwig, D. Gade, A. Beck, K. Borzym, K. Heitmann, R. Rabus, H. Schlesner, R. Amann, and R. Reinhardt. 2003. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl. Acad. Sci. USA 1008298-8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez, M. J., I. Cases, and A. Valencia. 2004. Gene order in prokaryotes: conservation and implications, p. 209-237. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in time and space. Kluwer Academic, New York, NY.
- 12.Griffiths, E., and R. S. Gupta. 2007. Phylogeny and shared conserved inserts in proteins provide evidence that Verrucomicrobia are the closest known free-living relatives of chlamydiae. Microbiology 1532648-2654. [DOI] [PubMed] [Google Scholar]
- 13.Hedlund, B. P., J. J. Gosink, and J. T. Staley. 1997. Verrucomicrobia div. nov., a new division of the bacteria containing three new species of Prosthecobacter. Antonie van Leeuwenhoek 7229-38. [DOI] [PubMed] [Google Scholar]
- 14.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42182-192. [Google Scholar]
- 15.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H. W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304728-730. [DOI] [PubMed] [Google Scholar]
- 16.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1804765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins, C., and J. A. Fuerst. 2001. Phylogenetic analysis of evolutionary relationships of the planctomycete division of the domain bacteria based on amino acid sequences of elongation factor Tu. J. Mol. Evol. 52405-418. [DOI] [PubMed] [Google Scholar]
- 18.Jenkins, C., R. Samudrala, I. Anderson, B. P. Hedlund, G. Petroni, N. Michailova, N. Pinel, R. Overbeek, G. Rosati, and J. T. Staley. 2002. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc. Natl. Acad. Sci. USA 9917049-17054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 20.Koch, A. L. 2003. Were Gram-positive rods the first bacteria? Trends Microbiol. 11166-170. [DOI] [PubMed] [Google Scholar]
- 21.Larsen, R. A., C. Cusumano, A. Fujioka, G. Lim-Fong, P. Patterson, and J. Pogliano. 2007. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 211340-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liesack, W., R. Soller, T. Steward, H. Haas, S. Giovannoni, and E. Stackebrandt. 1992. The influence of tachytelically (rapidly) evolving sequences on the topology of phylogenetic trees—intrafamily relationships and the phylogenetic position of Planctomycetaceae as revealed by comparative analysis of 16S ribosomal RNA sequences. Syst. Appl. Microbiol. 15357-362. [Google Scholar]
- 23.Lowe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391203-206. [DOI] [PubMed] [Google Scholar]
- 24.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 321363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machida, M., K. Takechi, H. Sato, S. J. Chung, H. Kuroiwa, S. Takio, M. Seki, K. Shinozaki, T. Fujita, M. Hasebe, and H. Takano. 2006. Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss. Proc. Natl. Acad. Sci. USA 1036753-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margolin, W. 2004. The assembly of proteins at the cell division site, p. 79-102. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in time and space. Kluwer Academic, New York, NY.
- 28.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCoy, A. J., and A. T. Maurelli. 2006. Building the invisible wall: updating the chlamydial peptidoglycan anomaly. Trends Microbiol. 1470-77. [DOI] [PubMed] [Google Scholar]
- 30.Mingorance, J., and J. Tamames. 2004. The bacterial dcw gene cluster: an island in the genome?, p. 249-271. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in Time and Space. Kluwer Academic, New York, NY.
- 31.Mingorance, J., J. Tamames, and M. Vicente. 2004. Genomic channeling in bacterial cell division. J. Mol. Recognit. 17481-487. [DOI] [PubMed] [Google Scholar]
- 32.Muller, T., and M. Vingron. 2000. Modeling amino acid replacement. J. Comput. Biol. 7761-776. [DOI] [PubMed] [Google Scholar]
- 33.Nikolaichik, Y. A., and W. D. Donachie. 2000. Conservation of gene order amongst cell wall and cell division genes in Eubacteria, and ribosomal genes in Eubacteria and eukaryotic organelles. Genetica 1081-7. [DOI] [PubMed] [Google Scholar]
- 34.Nogales, E., K. H. Downing, L. A. Amos, and J. Lowe. 1998. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol. 5451-458. [DOI] [PubMed] [Google Scholar]
- 35.Pilhofer, M., A. P. Bauer, M. Schrallhammer, L. Richter, W. Ludwig, K. H. Schleifer, and G. Petroni. 2007. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res. 35e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilhofer, M., G. Rosati, W. Ludwig, K. H. Schleifer, and G. Petroni. 2007. Coexistence of tubulins and ftsZ in different Prosthecobacter species. Mol. Biol. Evol. 241439-1442. [DOI] [PubMed] [Google Scholar]
- 37.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. Deboy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 281397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 312134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose, T. M., J. G. Henikoff, and S. Henikoff. 2003. CODEHOP (COnsensus-DEgenerate hybrid oligonucleotide primer) PCR primer design. Nucleic Acids Res. 313763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, T. M., E. R. Schultz, J. G. Henikoff, S. Pietrokovski, C. M. McCallum, and S. Henikoff. 1998. Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 261628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4654-666. [DOI] [PubMed] [Google Scholar]
- 42.Sanchez-Pulido, L., D. Devos, S. Genevrois, M. Vicente, and A. Valencia. 2003. POTRA: a conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci. 28523-526. [DOI] [PubMed] [Google Scholar]
- 43.Sangwan, P., X. L. Chen, P. Hugenholtz, and P. H. Janssen. 2004. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl. Environ. Microbiol. 705875-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sangwan, P., S. Kovac, K. E. Davis, M. Sait, and P. H. Janssen. 2005. Detection and cultivation of soil verrucomicrobia. Appl. Environ. Microbiol. 718402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18502-504. [DOI] [PubMed] [Google Scholar]
- 46.Strous, M., E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Medigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, V., B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H. W. Mewes, J. Weissenbach, M. S. Jetten, M. Wagner, and D. Le Paslier. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440790-794. [DOI] [PubMed] [Google Scholar]
- 47.Tamames, J. 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2RESEARCH0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamames, J., M. Gonzalez-Moreno, J. Mingorance, A. Valencia, and M. Vicente. 2001. Bringing gene order into bacterial shape. Trends Genet. 17124-126. [DOI] [PubMed] [Google Scholar]
- 49.Teeling, H., T. Lombardot, M. Bauer, W. Ludwig, and F. O. Glockner. 2004. Evaluation of the phylogenetic position of the planctomycete ‘Rhodopirellula baltica’ SH 1 by means of concatenated ribosomal protein sequences, DNA-directed RNA polymerase subunit sequences and whole genome trees. Int. J. Syst. Evol. Microbiol. 54791-801. [DOI] [PubMed] [Google Scholar]
- 50.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson, N. R., C. Yeats, K. Bell, M. T. Holden, S. D. Bentley, M. Livingstone, A. M. Cerdeno-Tarraga, B. Harris, J. Doggett, D. Ormond, K. Mungall, K. Clarke, T. Feltwell, Z. Hance, M. Sanders, M. A. Quail, C. Price, B. G. Barrell, J. Parkhill, and D. Longbottom. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Heijenoort, J. 1996. Murein synthesis, p. 1025-1034. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. Brooks Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schachter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 53.Vaughan, S., B. Wickstead, K. Gull, and S. G. Addinall. 2004. Molecular evolution of FtsZ protein sequences encoded within the genomes of archaea, bacteria, and eukaryota. J. Mol. Evol. 5819-39. [DOI] [PubMed] [Google Scholar]
- 54.Vicente, M., J. Alvarez, and R. Martinez-Arteaga. 2004. How similar cell division genes are located and behave in different bacteria, p. 239-248. In M. Vicente, J. Tamames, A. Valencia, and J. Mingorance (ed.), Molecules in Time and Space. Kluwer Academic, New York.
- 55.Vicente, M., M. J. Gomez, and J. A. Ayala. 1998. Regulation of transcription of cell division genes in the Escherichia coli dcw cluster. Cell Mol. Life Sci. 54317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vicente, M., and A. I. Rico. 2006. The order of the ring: assembly of Escherichia coli cell division components. Mol. Microbiol. 615-8. [DOI] [PubMed] [Google Scholar]
- 57.Vicente, M., A. I. Rico, R. Martinez-Arteaga, and J. Mingorance. 2006. Septum enlightenment: assembly of bacterial division proteins. J. Bacteriol. 18819-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner, M., and M. Horn. 2006. The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol. 17241-249. [DOI] [PubMed] [Google Scholar]
- 59.Ward, N. L., F. A. Rainey, B. P. Hedlund, J. T. Staley, W. Ludwig, and E. Stackebrandt. 2000. Comparative phylogenetic analyses of members of the order Planctomycetales and the division Verrucomicrobia: 23S rRNA gene sequence analysis supports the 16S rRNA gene sequence-derived phylogeny. Int. J. Syst. Evol. Microbiol. 501965-1972. [DOI] [PubMed] [Google Scholar]
- 60.Ward-Rainey, N., F. A. Rainey, and E. Stackebrandt. 1997. The presence of a dnaK (HSP70) multigene family in members of the orders Planctomycetales and Verrucomicrobiales. J. Bacteriol. 1796360-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weisburg, W. G., T. P. Hatch, and C. R. Woese. 1986. Eubacterial origin of chlamydiae. J. Bacteriol. 167570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wheeler, D. L., D. M. Church, S. Federhen, A. E. Lash, T. L. Madden, J. U. Pontius, G. D. Schuler, L. M. Schriml, E. Sequeira, T. A. Tatusova, and L. Wagner. 2003. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 3128-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wisotzkey, J. D., P. Jurtshuk, Jr., and G. E. Fox. 1990. PCR amplification of 16S rDNA from lyophilized cell cultures facilitates studies in molecular systematics. Curr. Microbiol. 21325-327. [DOI] [PubMed] [Google Scholar]
- 64.Yee, B., F. F. Lafi, B. Oakley, J. T. Staley, and J. A. Fuerst. 2007. A canonical FtsZ protein in Verrucomicrobium spinosum, a member of the bacterial phylum Verrucomicrobia that also includes tubulin-producing Prosthecobacter species. BMC Evol. Biol. 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.