Abstract
The psbA gene of the chloroplast genome has a codon usage that is unusual for plant chloroplast genes. In the present study the evolutionary status of this codon usage is tested by reconstructing putative ancestral psbA sequences to determine the pattern of change in codon bias during angiosperm divergence. It is shown that the codon biases of the ancestral genes are much stronger than all extant flowering plant psbA genes. This is related to previous work that demonstrated a significant increase in synonymous substitution in psbA relative to other chloroplast genes. It is suggested, based on the two lines of evidence, that the codon bias of this gene currently is not being maintained by selection. Rather, the atypical codon bias simply may be a remnant of an ancestral codon bias that now is being degraded by the mutation bias of the chloroplast genome, in other words, that the psbA gene is not at equilibrium. A model for the evolution of selective pressure on the codon usage of plant chloroplast genes is discussed.
Keywords: chloroplast, molecular evolution, selection
The nonuniform use of synonymous codons, or codon bias, that is observed in essentially all coding sequences is determined by an interaction of two main forces, composition bias and selection (1). To some degree the nucleotide bias observed in degenerate positions of coding sequences is the product of an overall genome compositional bias (2) and not a function of selection on coding sequences specifically. In some genomes, this composition bias is complemented by natural selection that acts to increase the frequency of specific codons (3), frequently resulting in a nucleotide bias at degenerate coding positions that is markedly different than the bias in noncoding sequences. Studies on codon bias have, for the most part, focused on determining the relative importance of these two factors in different organisms (1).
Because selective differences between codons are quite small (4), selection appears to be a factor only in those organisms, such as bacteria, which have a fairly large effective population size (5). The best example of an organism for which good evidence of selection exists is Escherichia coli, which provides a basic model for how selection affects codon bias. The relative frequency of synonymous codons in E. coli corresponds to the relative abundancies of the isoacceptor tRNAs in the cell, suggesting that selection has adapted codon usage to the tRNA population to increase the translation efficiency (3). This is supported by the observation that the degree of codon bias in different genes is correlated with expression level, such that highly expressed genes tend to have a much stronger bias in codon usage (5). Evidence supporting selection also comes from analyses of substitution rate, which show an inverse correlation between codon bias and synonymous substitution rate, suggesting that highly expressed genes in E. coli have stronger constraints on their codon usage (5, 6).
In our work we have been studying the codon bias of plant chloroplast genes. The chloroplast genome of plants contains approximately 100 genes, most of which code for proteins involved in either protein synthesis, including 31 tRNA genes, or photosynthesis (7). In terms of nucleotide composition, the genome has a high A+T content both in noncoding sequences and in degenerate positions of coding sequences (8), suggesting that codon bias is primarily a function of composition bias. The single exception is psbA, which codes for the core protein of photosystem II and has a noticeably different codon bias. This difference is most obvious in, but not limited to, the 2-fold degenerate groups with a pyrimidine at the third position (referred to here as NNY). At the third codon position of these groups, the psbA gene has a bias toward C unlike all other genes, which are strongly biased toward T (8). What makes this interesting is that only a single tRNA is available in the chloroplast to translate each of these synonymous groups. In each case this tRNA is complementary to the codon with a C at the third position, indicating that the psbA codon usage may be adapted to the chloroplast tRNA population (8). In fact, a close look at the unusual codon bias of the plant psbA gene shows that each synonymous group has a bias toward that codon that has an intermediate interaction strength with the available tRNA (9). Because the psbA gene has the highest level of translation among chloroplast genes (10), it has been proposed that the atypical codon usage of this gene is an adaptation to enhance translation efficiency (8, 9).
It also has been observed that the psbA gene has the lowest rate of synonymous substitution of all plant chloroplast genes, which supports the notion that selection is acting on the codon usage of psbA (11). However, this simple model has not held up under subsequent analysis. When synonymous substitution rate was measured separately for different synonymous groups, the results were exactly opposite what was expected if selection is constraining the unusual codon bias of the psbA gene. Those groups in which the codon usage of psbA is most atypical actually have a significantly increased rate of substitution in this gene relative to all other chloroplast genes (12). This observation is inconsistent with a model of constraining selection and has raised new questions concerning the role of selection in relation to the unusual codon usage of psbA.
In an attempt to explain both the unusual codon bias and the unexpected pattern of synonymous substitution rate in the psbA gene, two hypotheses have been put forward (12). The first hypothesis posits that the plant psbA gene is actually under positive selection, which currently is adapting the codon usage toward an even more strongly atypical pattern to increase translation efficiency. The increased rate of substitution in certain synonymous groups then would be analogous to increased rates of amino acid replacement under positive selection (13). The alternative hypothesis proposed is that the psbA gene was historically under constraining selection but that this selection recently has been relaxed. What is common to the two models is that both are based on the premise that the plant psbA gene is not currently at equilibrium. Rather, the difference in codon bias between psbA and the other chloroplast genes is either a reflection of an ongoing process of directional selection or simply a remnant of an ancestral codon bias that is degrading (12).
In the current work we have tested these two models based on the fact that they generate two different predictions concerning the evolution of codon bias at the psbA locus during the divergence of the flowering plants. If positive selection has been acting, the psbA gene will have been evolving toward a more atypical codon bias pattern. In contrast, under the second hypothesis, in which selection recently was relaxed, the codon bias of the psbA gene has been evolving toward the equilibrium composition bias, becoming more similar to the other chloroplast genes over time.
The sequence of the ancestral flowering plant psbA gene was reconstructed, and the codon bias of the putative ancestral and extant psbA genes was compared using the codon adaptation index (CAI) of reference (6). As a measure of codon bias, CAI shows the degree of adaptation to a specific pattern of codon usage, and in this case we measured it relative to the atypical codon bias pattern. Therefore, the relative CAI values of different genes will indicate the degree to which they are adapted to the pattern of codon usage observed in the psbA gene. Under the first hypothesis, the expectation is that the CAI value of the ancestral sequence would be lower than the CAI value of all angiosperm psbA genes, indicating that adaptation has been increasing over time. Alternatively, under the second model, the CAI value of the ancestral psbA gene is expected to be greater than the CAI value of all angiosperm psbA genes. The results are shown to strongly support the second model. Taken together with the previous rate results (12) the present study is consistent with the proposal that the atypical codon bias of the psbA gene is a remnant of an ancestral codon bias that is not currently being constrained by selection. This possibility raises interesting questions concerning the evolution and study of codon bias.
MATERIALS AND METHODS
All available psbA sequences from flowering plants were extracted from GenBank as listed in Table 1. Because the only monocot sequences available were from members of the grass family, DNA was extracted from Phoenix dactylifera, Acorus gramineus, and Ludisia discolor following the method of Doyle and Doyle (14). The psbA gene was amplified from these species using two sets of primers. The first amplification used primers 1F (GCTAGGTCTAGAGGGAAGTTGTGAG) and 980R (CGTTCGTGCATTACTTCCATAC), and the second used primers 2F (GGGGTCGCTTCTGCAACTG) and 1000R (GCTAGGTCTAGAGGGAAGTTGTGAG). Both amplifications involved 40 cycles of 95°C for 1 min, 42°C for 1 min, and 72°C for 1 min. The PCR products were sequenced directly after a previously described procedure (15) using internal primers designed directly from the sequences obtained.
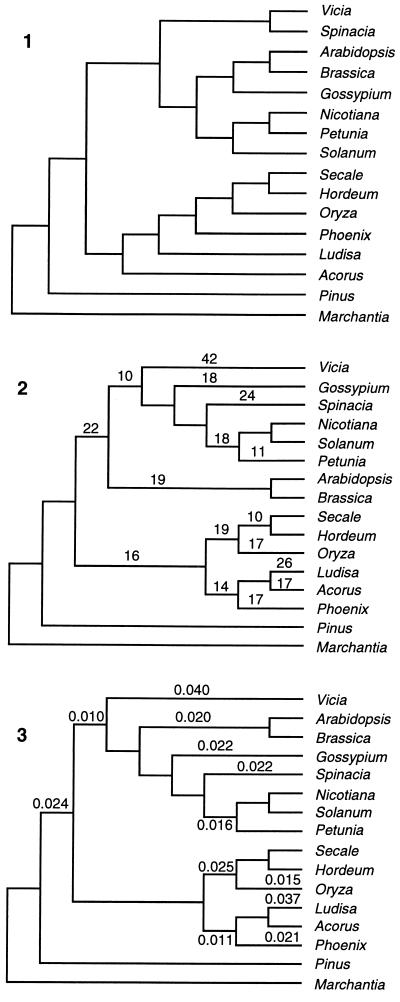
The character trace function of McClade 3.0 (16) was used to reconstruct the ancestral sequence. Because of the uncertainty about the relationships between the families of plants represented, three different topologies (Fig. 1) were used to generate three putative ancestral sequences (Anc1, Anc2, and Anc3). The first topology is based on a published classification (17), the second is the topology generated by parsimony analysis using paup (18) with equal character weighting, and the third is the topology generated using neighbor joining (19) with Kimura 2-parameter distances and a Ts/Tv of 2:1 by the phylip package (20). In all three, Pinus and Marchantia polymorpha were used as outgroups (Fig. 1). The ancestral sequence was taken as the sequence inferred at the point of divergence of the monot and dicot sequences. The 20 sites that were ambiguous (1.9% of the sequence) were excluded from the analysis.
Figure 1.
The three topologies used to generate the different potential ancestral genes. The source of each topology is given in the text. For topology 2, generated by maximum parsimony analysis, the number of substitutions is given for each branch with 10 or more inferred substitutions. For topology 3, generated by neighbor joining, branch lengths are given for branches with 0.010 substitutions per site or more. Neither topology is drawn to scale. The labels 1, 2, and 3 indicate the topologies used to generate genes Anc1, Anc2, and Anc3, respectively.
For each gene the CAI was calculated by the formula from ref. 6. To calculate CAI, each codon is assigned a fitness, which is calculated from a reference codon usage pattern. The relative frequencies of synonymous codons are taken as the fitness values so that the highly represented codon in a synonymous group is assigned a fitness of 1 and all other fitness values range from 0 to 1. As a result, a CAI that is calculated based on these fitness values is a measure of the tendency to use high fitness codons, that is, a measure of adaptation to the codon usage pattern used as the reference. For every gene in this analysis, CAI was calculated relative to several different reference psbA genes—the three reconstructed ancestral genes, the tobacco and rice psbA genes, the psbA gene from M. polymorpha, and the psbA gene from the alga Chlamydomonas reinhardtii. The C. reinhardtii gene has a very strong codon bias with a pattern identical to the plant psbA gene (8, 9). Therefore, CAI values represent adaptation to the codon usage pattern that appears to arise from selection.
RESULTS AND DISCUSSION
The Change in Codon Bias of psbA During Flowering Plant Divergence.
The two hypotheses concerning the evolution of codon bias at the psbA locus (12) lead to opposing predictions about the pattern of change in the codon bias during angiosperm divergence. In this study we attempted to eliminate one of the two by comparing the CAI values of reconstructed ancestral sequences to the CAI of existing genes to determine the pattern of change over time. Three possible ancestral flowering plant psbA genes (Anc1, Anc2, and Anc3) were reconstructed, one for each of the topologies in Fig. 1, and the CAI values based on several reference genes (Table 2) indicate a definite evolutionary trend. Regardless of which gene with an atypical codon bias is used as the basis, the three ancestral sequences have higher CAI values than all extant genes. Only a single minor exception to this general trend is observed. When the Oryza sativa psbA gene is the reference, which is the reference with the weakest atypical pattern, the CAI of the extant Secale gene has a CAI slightly greater than Anc1. Despite this exception, a comparison of CAI values shows that the putative ancestral psbA genes all have a codon bias that is more strongly adapted to the atypical codon bias than do the extant psbA genes.
The same general trend also is observed if we compare the composition of third codon positions directly. Two main sets of synonymous codon groups result in which psbA is most noticeably unusual in its codon bias, the NNY 2-fold degenerate groups and the 4-fold degenerate groups (9). For both of these sets, the general pattern of evolution is away from the bias observed in psbA (Table 3). For the NNY 2-fold degenerate synonymous groups, in which psbA is biased toward C at the third codon position, the direction of evolution is predominantly, although not exclusively, toward a lower C content over angiosperm divergence. Similarly, in the 4-fold degenerate synonymous groups, psbA has an unusually high T content at the third codon position, and extant genes have a lower T content than the putative ancestral sequences. Therefore, the evolutionary trend indicated by CAI is consistent with the trend observed at the nucleotide level. Overall, the results provide strong evidence that the psbA gene is evolving predominantly away from the atypical codon pattern.
Selection and the Codon Usage of psbA.
Although the plant psbA gene has a codon bias pattern that is different than all other plant chloroplast genes, recent work has raised questions concerning the role of selection in generating this atypical pattern. On the one hand, it seems clear that selection must play some role (9, 12). At the third position of specific codon groups, a much higher frequency of C is observed in psbA (over 60% in the case of tobacco, see Table 3) than in either noncoding DNA (16% in the tobacco genome) or the same synonymous groups of any other gene (8). On its own, the unique base frequency is indicative of selection, but the match of psbA codon bias to the tRNA population (9) as well as the very high translation level of the gene due to rapid turnover (10) make it a very strong case.
In spite of these observations concerning composition, the role of selection is drawn into question by the evolutionary dynamics of psbA. First, the NNY codon groups for which psbA has an atypical codon usage also have significantly increased rates of synonymous substitution relative to both the same synonymous groups in other genes as well as other synonymous groups in psbA (12). Second, the results from this study indicate that the degree to which psbA is atypical in its codon bias has been decreasing during angiosperm evolution (Tables 2 and 3). Both of these observations are inconsistent with selective constraints on the codon usage of psbA so alternative explanations are required. Of the two original hypotheses put forward to explain the evolution of codon bias in psbA (12) the one involving positive selection now can be eliminated. The general decrease that is observed in the degree of adaptation to the atypical codon bias pattern is not consistent at all with positive selection currently adapting the codon bias of the psbA gene toward a more strongly atypical pattern, which would be supported only if CAI consistently increased from ancestral to extant genes.
On the other hand, none of the unusual features of psbA evolution are inconsistent with the second model (12)—that selection on psbA recently has been relaxed so that the atypical codon bias of this gene is a remnant of an ancestral codon bias that is not being maintained presently in the flowering plants. The features that are inconsistent either with positive selection or with constraining selection can be explained under this model. First, the atypical codon bias would simply result from psbA not being at compositional equilibrium. Second, the pattern of substitution rates would be a product of the unusual base composition of psbA coupled with the assymetrical substitution process in the chloroplast genome (12). Finally, the consistent decrease in CAI would be a result of the degradation of the ancestral codon bias by a mutation bias that is no longer countered by selection. Instead, the current evolution of psbA appears to be dominated by whatever mechanism is driving the A+T bias in all other chloroplast genes and noncoding sequences. The consistency between the pattern of evolution in psbA and this model, but not other models, requires that it be seriously considered.
A Model for the Evolution of Codon Bias in Chloroplast Genes.
The basic proposal, that selection on psbA has been recently relaxed, now can be examined in further detail and extended to all chloroplast genes. If the codon bias of psbA is actually an historic remnant, then it means that selective pressure has shifted in the recent past. However, this does not require a change in selection on codon bias itself, rather it seems more likely that the relaxation of selection was actually an epiphenomenon. The original proposal (12) was that a relaxation of selection could have been linked to changes in genome copy number. Because the plant chloroplast contains a very large number of identical genomes, unlike the single genome in some algae, such as C. reinhardtii (21), selection for translation efficiency of individual transcripts can be replaced by the number of transcripts available, as long as the overall translation rate is sufficient to maintain protein levels. If genome copy number gradually has increased over time then we can explain the unique nature of psbA by the fact that selection on codon usage would be relaxed at different times for different genes, persisting longest in the highly expressed genes. This also could explain the slight correlation between codon usage and expression level in plant chloroplast genes, including the relatively high CAI of the highly expressed rbcL gene (11).
To this it should be added that a changing effective population size would have a very similar effect. The selective advantage of an optimal codon is likely to be quite small, the one measurement that has been made is on the order of 10−9 for the gnd locus in E. coli (4), so that selection on codon usage can overcome drift only if the population size is quite large (5). If effective population size has been decreasing as multicellular higher plants have evolved from a unicellular ancestor, a specific point in time will exist at which selective differences are no longer large enough to overcome drift. Because selection intensity varies among genes, this point in time will be different for each gene. Again, highly expressed genes would have had a codon bias determined largely by selection until much later in plant evolution.
Although this model is consistent with the unusual compositional and evolutionary properties of psbA, certainly more consistent than the alternatives, several features remain to be examined further. One issue that needs to be addressed is the development of expectations about the evolution of CAI and composition in the absence of selection. This would be particularly useful for assessing the variation in CAI among extant psbA genes that is observed (Table 2), which may be random noise but also could arise from differences, such as variation in population size, among lineages. It also could provide an expected time frame for degeneration of codon bias for a comparison of the observed change in CAI to an estimate based on divergence times for the monocots and dicots (22). Given the complex dynamics of context dependency that have been observed for base substitutions in chloroplast DNA (23, 24) this will not be a simple matter if we are to generate an accurate expectation. Finally, it will be interesting to analyze other genes, specifically genes suspected to be under no selection for codon usage. Because the codon bias of rbcL is also slightly atypical (11), this gene, although sequenced for many plants, is not a good candidate. Finally, it should be emphasized that this model does not require that selection has been completely relaxed in psbA, only that the intensity has decreased recently nor that selection intensity is equal across different lineages.
Reconstruction of Ancestral Sequences.
Because the test used here is based on the reconstruction of an ancestral sequence we also must consider the reliability of this approach. First, the reconstruction of ancestral sequences is a function of topology. To account for this, three different topologies were used, and because the different topologies give essentially identical answers, topology does not appear to be a factor in this particular analysis.
Two other potential difficulties must be addressed. The first is the reliability of the ancestral sequence given the variation among extant sequences. Basically, the further the sequences have diverged the more problematic it is to infer the ancestral sequence due to multiple substitutions per site. This has been confirmed by simulation analyses, which have shown that reconstruction is extremely reliable for closely related sequences, particularly at divergence levels below 0.1 substitutions per site (25). This high reliability is true for both parsimony, used here, and likelihood methods (26), and at low levels of divergence all methods of reconstruction are equally reliable (26). Given these observations it is expected that our reconstruction is very reliable. The psbA gene is highly conserved, and branch lengths for the neighbor-joining tree (Fig. 1) shows that the longest branch is 0.042 substitutions per site, and most are less than 0.025. For the parsimony tree, branch lengths also are given. The psbA gene is 1,056 nucleotides in length, excluding the start and stop codons, and the longest branch has only 42 substitutions (0.040 per site). In addition, a distribution of the minimum number of changes per site (Table 4) shows that, of the total of 1,056 sites, 849 are conserved among all flowering plants in this study. Therefore, even if all changes are assumed to occur at the third position, 142 of 353 codons are fully conserved. Overall, very few sites have more than two inferred changes over all species, and most variable sites have a single change. All of these lines of evidence indicate extremely low levels of divergence, well below the level at which reconstruction is found to be highly reliable. Therefore, it is very unlikely that errors in reconstruction could generate the overwhelming trend observed in Tables 2 and 3.
The second issue is how selection might affect the reconstruction. One factor that can confound this method is convergent evolution, and the likelihood of this increases with adaptive selection. Although this can strongly affect certain types of analyses, it was not considered a problem for this analysis because of the nature of selection on codon usage. Convergent evolution will affect reconstructions when strong selection for a specific character is at a given site, for instance when a certain amino acid is favored at a location in a protein so that different lineages will converge on that amino acid. However, selection on codon bias is not limited to specific sites. Instead, a mutation at any number of sites will increase translation efficiency so that it is unlikely that convergence in a large number of different lineages has occurred at a significant number of sites. Therefore, it seems very unlikely that the results are an artifact of such selection.
Conclusions.
The codon bias of plant chloroplast genes is proving to be a much more interesting and complicated issue than first supposed (27) as it is now clear that both the codon bias and the evolutionary dynamics of the psbA gene are quite unusual. These unusual features are not consistent with constraining selection, forcing us to consider models in which the psbA gene is not at equilibrium (12). In the current study we have provided strong evidence that positive selection is not responsible. However, the results of this analysis, as well as previous studies (11, 12), all are consistent with the hypothesis that the unusual codon bias of the psbA gene is not currently being maintained by selection. It is proposed that selection on this gene recently has been relaxed so that the atypical codon bias of this gene is actually a remnant of this ancestral bias that is degrading toward the compositional bias. Given the many complexities of this issue it remains far from resolved, but the data clearly show that the plant psbA gene provides an excellent opportunity to investigate an unusual evolutionary situation. It also leads to predictions concerning other lineages in that a relationship between genome number and/or population size and selection is postulated. Further testing of this hypothesis, therefore, will be possible with the accumulation of further data.
Table 1.
Accessions for the psbA sequences used in this study
| Species | Accession number* |
|---|---|
| Monocots | |
| Secale cereale | X13327 |
| Oryza sativa | M36191 |
| Hordeum vulgare | X07942 |
| Acorus gramineus | This study |
| Phoenix dactylifera | This study |
| Ludisia discolor | This study |
| Dicots | |
| Petunia hybrida | X04974 |
| Nicotiana debneyi | J01448 |
| Solanum nigrum | U25659 |
| Spinacia oleracea | J01442 |
| Vicia faba | X17694 |
| Arabidopsis thaliania | X79898 |
| Brassica napus | M36720 |
| Gossypium hisutum | X15885 |
| Other | |
| Pinus contorta | X53721 |
| Marchantia polymorpha | X04465 |
| Chlamydomonas rheinhardtii | X01424 |
GenBank accession number except for those species sequenced in this analysis.
Table 2.
Comparison of CAI values from reconstructed ancestral and extant psbA sequences
| Species | Reference psbA gene*
|
||||||
|---|---|---|---|---|---|---|---|
| Anc1 | Anc2 | Anc3 | Cre | Mpo | Osa | Nta | |
| Ancestral | |||||||
| Anc1 | 0.723 | 0.714 | 0.710 | 0.375 | 0.592 | 0.737 | 0.747 |
| Anc2 | 0.744 | 0.739 | 0.734 | 0.385 | 0.624 | 0.759 | 0.764 |
| Anc3 | 0.742 | 0.737 | 0.733 | 0.378 | 0.617 | 0.755 | 0.762 |
| Monocots | |||||||
| Hordeum | 0.646 | 0.643 | 0.638 | 0.302 | 0.552 | 0.709 | 0.687 |
| Secale | 0.680 | 0.674 | 0.668 | 0.348 | 0.577 | 0.741 | 0.720 |
| Oryza | 0.649 | 0.641 | 0.637 | 0.318 | 0.538 | 0.710 | 0.690 |
| Acorus | 0.664 | 0.649 | 0.644 | 0.319 | 0.527 | 0.684 | 0.711 |
| Phoenix | 0.684 | 0.678 | 0.670 | 0.333 | 0.565 | 0.721 | 0.730 |
| Ludisia | 0.628 | 0.615 | 0.610 | 0.278 | 0.483 | 0.660 | 0.662 |
| Dicots | |||||||
| Arabidopsis | 0.663 | 0.660 | 0.653 | 0.319 | 0.546 | 0.697 | 0.712 |
| Brassica | 0.684 | 0.679 | 0.675 | 0.346 | 0.573 | 0.713 | 0.726 |
| Gossypium | 0.650 | 0.641 | 0.638 | 0.325 | 0.551 | 0.699 | 0.710 |
| Nicotiana | 0.636 | 0.622 | 0.619 | 0.337 | 0.525 | 0.666 | 0.704 |
| Petunia | 0.638 | 0.626 | 0.623 | 0.336 | 0.524 | 0.669 | 0.701 |
| Solanum | 0.628 | 0.613 | 0.610 | 0.335 | 0.522 | 0.660 | 0.695 |
| Spinacia | 0.662 | 0.649 | 0.650 | 0.319 | 0.538 | 0.696 | 0.710 |
| Vicia | 0.614 | 0.604 | 0.603 | 0.291 | 0.512 | 0.659 | 0.670 |
Reference gene for relative synonymous codon use values (see text). Cre, C. reinhardtii; Mpo, M. polymorpha; Osa, O. sativa, and Nta, N. tabacum. For each reference gene the extant psbA with the largest CAI is underlined.
Table 3.
Composition of reconstructed ancestral and extant psbA genes
| Species | %C 2-fold* | %T 4-fold† |
|---|---|---|
| Ancestral | ||
| Anc1 | 0.600 | 0.706 |
| Anc2 | 0.603 | 0.735 |
| Anc3 | 0.594 | 0.727 |
| Monocots | ||
| Hordeum | 0.390 | 0.683 |
| Secale | 0.571 | 0.689 |
| Oryza | 0.571 | 0.636 |
| Acorus | 0.559 | 0.660 |
| Phoenix | 0.578 | 0.637 |
| Ludisia | 0.483 | 0.645 |
| Dicots | ||
| Arabidopsis | 0.546 | 0.645 |
| Brassica | 0.558 | 0.680 |
| Gossypium | 0.546 | 0.612 |
| Nicotiana | 0.623 | 0.612 |
| Petunia | 0.610 | 0.622 |
| Solanum | 0.610 | 0.620 |
| Spinacia | 0.507 | 0.653 |
| Vicia | 0.506 | 0.590 |
C content of the NNY synonymous groups (see text).
T content of 4-fold degenerate synonymous groups.
Table 4.
Substitutions per site in psbA
| Number of substitutions | Number of sites
|
||
|---|---|---|---|
| Topology 1 | Topology 2 | Topology 3 | |
| 0 | 849 | 849 | 849 |
| 1 | 115 | 121 | 120 |
| 2 | 45 | 47 | 47 |
| 3 | 34 | 31 | 29 |
| 4 | 11 | 8 | 10 |
| 5 | 4 | 4 | 5 |
| 6 | 2 | 0 | 0 |
Acknowledgments
We would like to thank Michael Clegg, Brandon Gaut, and Gerald H. Learn for discussion of this work.
ABBREVIATION
- CAI
codon adaptation index
Footnotes
References
- 1.Sharp P M, Li W-H. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 2.Bernardi G, Bernardi G. J Mol Evol. 1986;24:1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- 3.Ikemura T. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 4.Hartl D L, Moriyama E N, Sawyer S. Genetics. 1994;138:227–234. doi: 10.1093/genetics/138.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W-H. J Mol Evol. 1987;24:337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- 6.Sharp P M, Li W-H. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer J D. In: Cell Culture and Somatic Genetics of Plants. Bogorad L, Vasil K, editors; Bogorad L, Vasil K, editors. 7A. New York: Academic; 1991. pp. 5–53. [Google Scholar]
- 8.Morton B R. J Mol Evol. 1993;37:273–280. doi: 10.1007/BF00175504. [DOI] [PubMed] [Google Scholar]
- 9.Morton B R. J Mol Evol. 1996;43:28–31. doi: 10.1007/BF02352296. [DOI] [PubMed] [Google Scholar]
- 10.Mullet J E, Klein R R. EMBO J. 1987;6:1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morton B R. Mol Biol Evol. 1994;11:231–238. doi: 10.1093/oxfordjournals.molbev.a040105. [DOI] [PubMed] [Google Scholar]
- 12.Morton B R. Mol Biol Evol. 1997;14:412–419. doi: 10.1093/oxfordjournals.molbev.a025777. [DOI] [PubMed] [Google Scholar]
- 13.Hughes A L, Nei M. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle J J, Doyle J L. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 15.Morton B R, Clegg M T. J Mol Evol. 1995;41:597–603. doi: 10.1007/BF00175818. [DOI] [PubMed] [Google Scholar]
- 16.Maddison W P, Maddison J R. MacClade: Analysis of Phylogeny and Character Evolution. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 17.Thorne R F. Bot Rev. 1992;58:225–348. [Google Scholar]
- 18.Swofford D. paup: Phylogenetic Analysis Using Parsimony, Version 3.1. Champaign, IL: Illinois Natural History Survey; 1993. [Google Scholar]
- 19.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. phylip, Version 3.5. Seattle: Univ. of Washington; 1992. [Google Scholar]
- 21.van den Hoek C. Algae: An Introduction to Phycology. New York: Cambridge Univ. Press; 1995. [Google Scholar]
- 22.Wolfe K H, Gouy M, Yang Y-W, Sharp P M, Li W-H. Proc Natl Acad Sci USA. 1989;86:6201–6205. doi: 10.1073/pnas.86.16.6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton B R. Proc Natl Acad Sci USA. 1995;92:9717–9721. doi: 10.1073/pnas.92.21.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morton B R. Mol Biol Evol. 1997;14:189–194. doi: 10.1093/oxfordjournals.molbev.a025777. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Nei M. J Mol Evol. 1997;44:S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z, Kumar S, Nei M. Genetics. 1995;141:1641–1650. doi: 10.1093/genetics/141.4.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfe K H, Sharp P M. Gene. 1988;66:215–222. doi: 10.1016/0378-1119(88)90358-7. [DOI] [PubMed] [Google Scholar]