Abstract
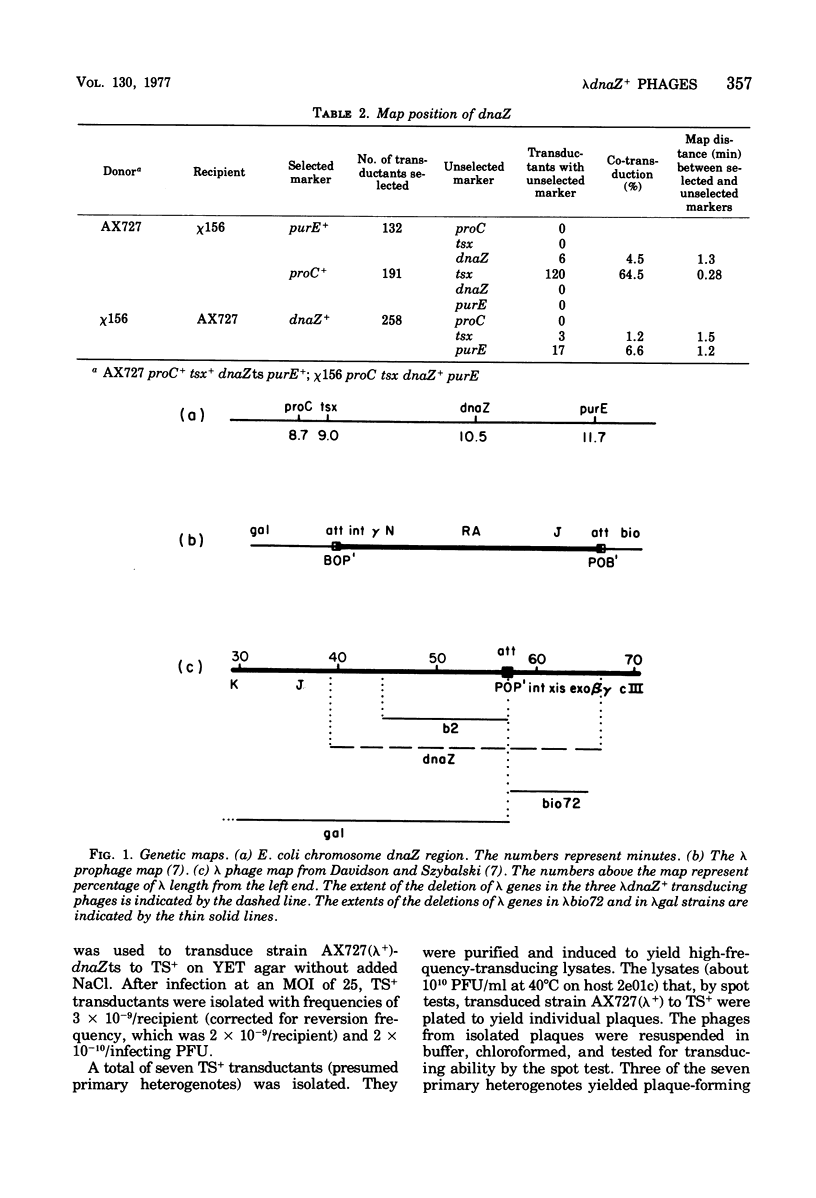
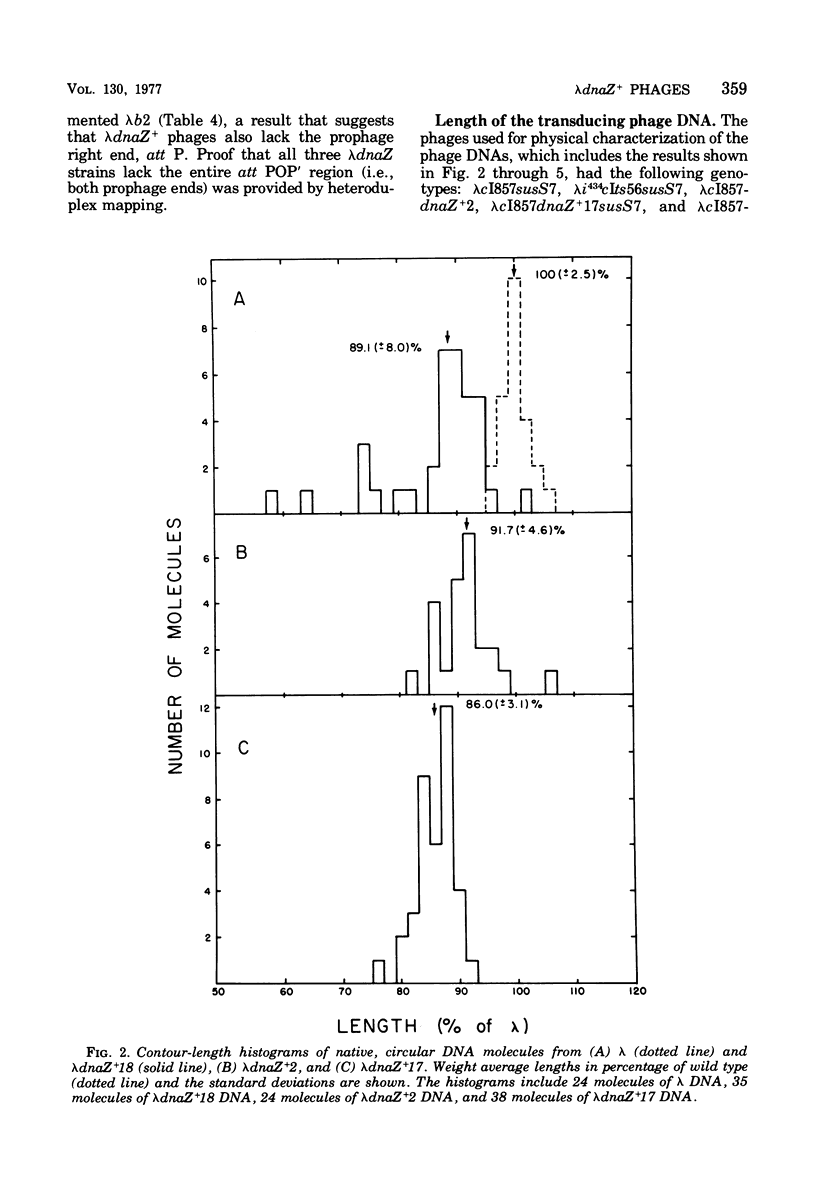
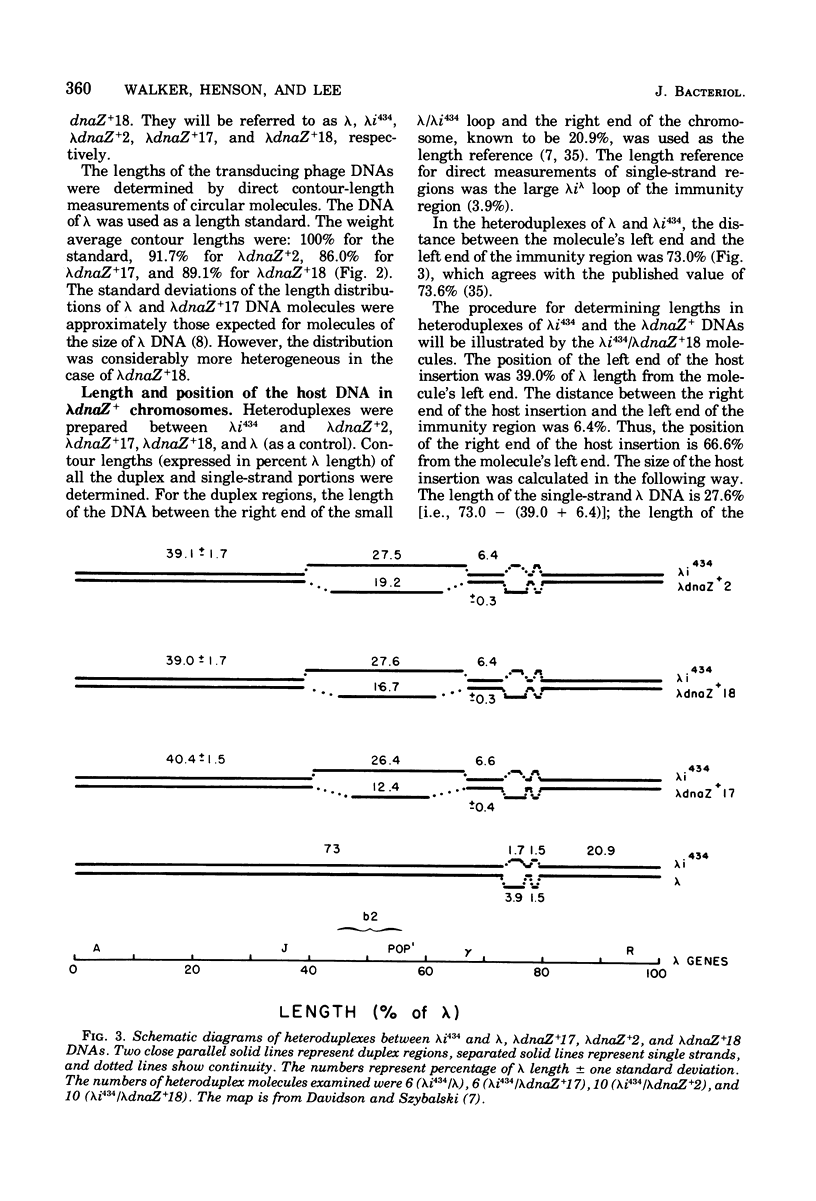
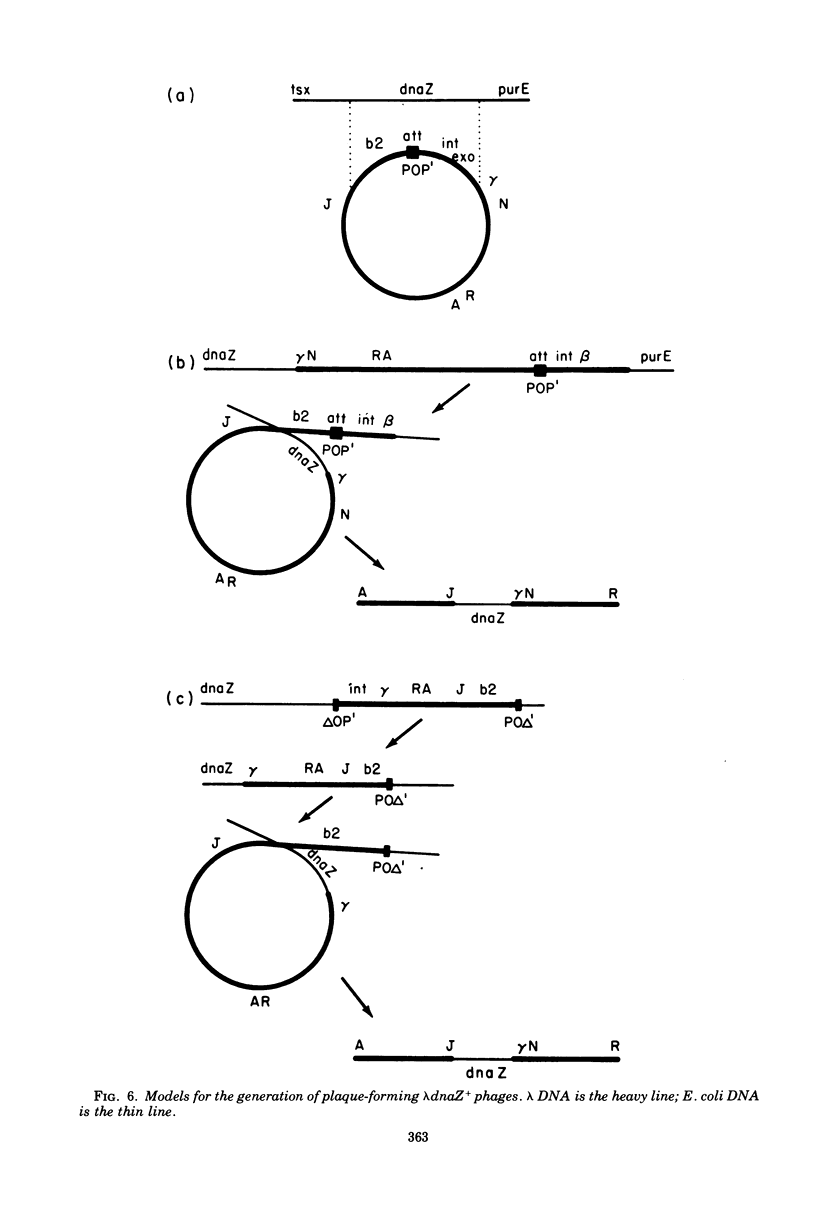
The Escherichia coli dnaZ gene, a deoxyribonucleic acid (DNA) polymerization gene, is located 1.2 min counterclockwise from purE, at approximately min 10.5 on the E. coli map. From a lysogen with lamdacI857 integrated at a secondary attachment site near purE, transducing phages (lambdadnaS+) that transduced a dnaZts (lambda+) recipient to temperature insensitivity (TS+) were discovered. Three different plaque-forming transducing phages were isolated from seven primary heterogenotes. Genetic tests and heteroduplex mapping were used to determine the length and position of E. coli DNA within the lambda DNA. Complementation tests demonstrated that the deletions in all three strains removed both att P and the int gene, i,e., DNA from both prophage ends. Heteroduplex mapping confirmed this result by demonstrating that all three strains had deletions of lambda DNA that covered the b2 to red region, thereby removing both prophage ends. Specifically, the deletions removed lambda DNA between the points 39.3 to 66.5% of lambda length (measured in percent length from the left and of lambda phage DNA) in all three strains. The three strains are distinct, however, because they had differing lengths of host DNA insertions. These phages must have been formed by an anomalous procedure, because standard lambda transducing phages are deleted for one prophage end only. In lambdagal and lambdabio strains, the deletions of lambda DNA begin at the union of prophage ends (i.e., position 57.3% of lambda length) and extend leftward or rightward, respectively (Davidson and Szybalski, in A, D. Hershey [ed.], The Bacteriophage Lambda, p. 45-82, 1971). Models for formation of the lambdadnaZ+ phages are discussed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom F. R., McFall E., Young M. C., Carothers A. M. Positive control in the D-serine deaminase system of Escherichia coli K-12. J Bacteriol. 1975 Mar;121(3):1092–1101. doi: 10.1128/jb.121.3.1092-1101.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMPBELL A. Transduction and segregation in Escherichia coli K12. Virology. 1957 Oct;4(2):366–384. doi: 10.1016/0042-6822(57)90070-3. [DOI] [PubMed] [Google Scholar]
- Fiandt M., Szybalski W., Malamy M. H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119(3):223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- Filip C. C., Allen J. S., Gustafson R. A., Allen R. G., Walker J. R. Bacterial cell division regulation: characterization of the dnaH locus of Escherichia coli. J Bacteriol. 1974 Aug;119(2):443–449. doi: 10.1128/jb.119.2.443-449.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingery R., Echols H. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1507–1514. doi: 10.1073/pnas.58.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. The integration and excision of the bacteriophage lambda genome. Cold Spring Harb Symp Quant Biol. 1968;33:735–747. doi: 10.1101/sqb.1968.033.01.084. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Rosner J. L. Acquisition of a determinant for chloramphenicol resistance by coliphage lambda. Proc Natl Acad Sci U S A. 1975 Dec;72(12):5041–5045. doi: 10.1073/pnas.72.12.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Beckwith J. R. Directed transposition of the arabinose operon: a technique for the isolation of specialized transducing bacteriophages for any Escherichia coli gene. J Mol Biol. 1969 Aug 28;44(1):117–127. doi: 10.1016/0022-2836(69)90408-2. [DOI] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang W. G., Walker J. R. Parental RF formation on phages phiX174 and M13 requires the dnaZ gene product of Escherichia coli. Biochem Biophys Res Commun. 1976 Jun 7;70(3):932–938. doi: 10.1016/0006-291x(76)90681-1. [DOI] [PubMed] [Google Scholar]
- Henderson D., Weil J. The nature and origin of a class of essential gene substitutions in bacteriophage lambda. Virology. 1975 Sep;67(1):124–135. doi: 10.1016/0042-6822(75)90410-9. [DOI] [PubMed] [Google Scholar]
- Hirsch H. J., Starlinger P., Brachet P. Two kinds of insertions in bacterial genes. Mol Gen Genet. 1972;119(3):191–206. doi: 10.1007/BF00333858. [DOI] [PubMed] [Google Scholar]
- Manly K. F. Biotin-transducing lambda phages with unusual att regions. Virology. 1970 Sep;42(1):138–143. doi: 10.1016/0042-6822(70)90246-1. [DOI] [PubMed] [Google Scholar]
- Manly K. F., Signer E. R., Radding C. M. Nonessential functions of bacteriophage lambda. Virology. 1969 Feb;37(2):177–188. doi: 10.1016/0042-6822(69)90197-4. [DOI] [PubMed] [Google Scholar]
- Neubauer Z. Prophage deletions selected by heat induction of bacteria lysogenic for lambda-int6CI857. Virology. 1970 Sep;42(1):225–228. doi: 10.1016/0042-6822(70)90257-6. [DOI] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Strathern A., Herskowitz I. Defective prophage in Escherichia coli K12 strains. Virology. 1975 Sep;67(1):136–143. doi: 10.1016/0042-6822(75)90411-0. [DOI] [PubMed] [Google Scholar]
- Szybalski E. H., Szybalski W. Physical mapping of the att-N region of coliphage lambda: apparent oversaturation of coding capacity in the gam-ral segment. Biochimie. 1974;56(11-12):1497–1503. doi: 10.1016/s0300-9084(75)80272-0. [DOI] [PubMed] [Google Scholar]
- Taketo A. Replication of phiA and phiX174 in Escherichia coli mutants thermosensitive in DNA synthesis. Mol Gen Genet. 1975 Sep 8;139(4):285–291. doi: 10.1007/BF00267968. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Truitt C. L., Walker J. R. Growth of phages lambda, phiX174, and Ml3 requires the dnaZ (previously dnaH) gene product of Escherichia coli. Biochem Biophys Res Commun. 1974 Dec 11;61(3):1036–1042. doi: 10.1016/0006-291x(74)90259-9. [DOI] [PubMed] [Google Scholar]
- Walker J. R., Ussery C. L., Allen J. S. Bacterial cell division regulation: lysogenization of conditional cell division lon - mutants of Escherichia coli by bacteriophage. J Bacteriol. 1973 Mar;113(3):1326–1332. doi: 10.1128/jb.113.3.1326-1332.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg R. A., Gottesman M. E. The integration and excision defect of bacteriophage lambda-dg. J Mol Biol. 1969 Dec 28;46(3):565–580. doi: 10.1016/0022-2836(69)90196-x. [DOI] [PubMed] [Google Scholar]
- Westmoreland B. C., Szybalski W., Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969 Mar 21;163(3873):1343–1348. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]
- Wickner S., Hurwitz J. Involvement of escherichia coli dnaZ gene product in DNA elongation in vitro. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1053–1057. doi: 10.1073/pnas.73.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S., Clark A. J., Low B. Genetic location of certain mutations conferring recombination deficiency in Escherichia coli. J Bacteriol. 1969 Jan;97(1):244–249. doi: 10.1128/jb.97.1.244-249.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zissler J. Integration-negative (int) mutants of phage lambda. Virology. 1967 Jan;31(1):189–189. doi: 10.1016/0042-6822(67)90030-x. [DOI] [PubMed] [Google Scholar]