Abstract
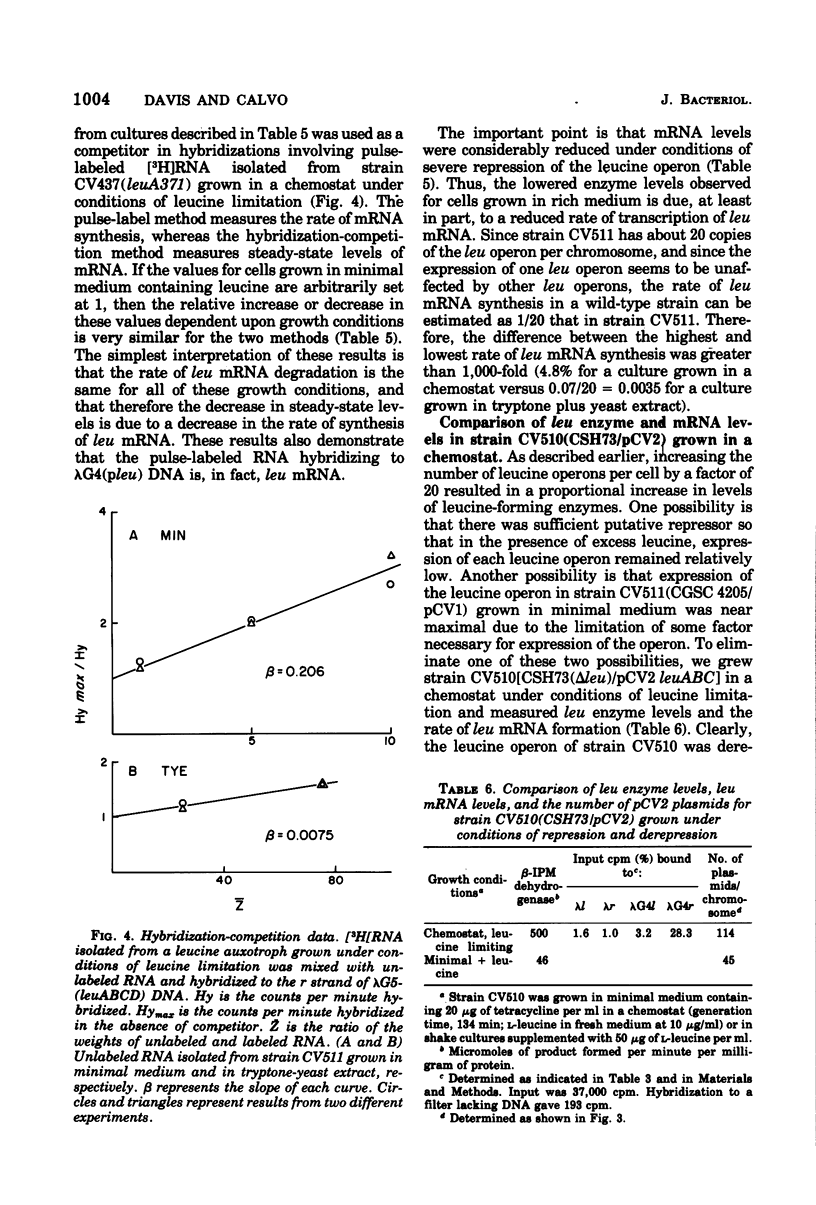
The levels of leucine-forming enzymes in Escherichia coli K-12 varied over a several thousand-fold range, depending upon conditions of growth. The highest levels were achieved by growing auxotrophs in a chemostat under conditions of leucine limitation. Under such conditions, enzyme levels were increased 45- to 90-fold relative to cells grown in minimal medium containing leucine (the latter values arbitrarily called 1). Leucine operon-specific messenger ribonucleic acid levels were elevated to about the same extent as enzyme levels in cells grown in a chemostat. Growth in media of greater complexity resulted in progressively lower levels of leucine-forming enzymes, reaching a value of less than 0.02 for growth in a medium containing tryptone broth and yeast extract. The levels of leucine operon-specified enzymes and messenger ribonucleic acid were also measured in strains containing about 25 copies of plasmid pCV1(ColE1-leu) per chromosome. For such strains grown in minimal medium, enzyme levels were proportional to the number of plasmids per cell. Furthermore, they followed the same trends as those described above upon derepression in a chemostat or upon repression following growth in rich media. Leucine messenger ribonucleic acid, measured both by pulse-labeling and hybridization-competition experiments, was roughly proportional to enzyme levels over this entire range. For a plasmid-containing strain grown in a chemostat under conditions of leucine limitation (about 100 plasmids per chromosome), about 27% of pulse-labeled ribonucleic acid was coded for by genes in or adjacent to the leucine operon, and 10% of the total protein was β-isopropylmalate dehydrogenase.
Full text
PDF


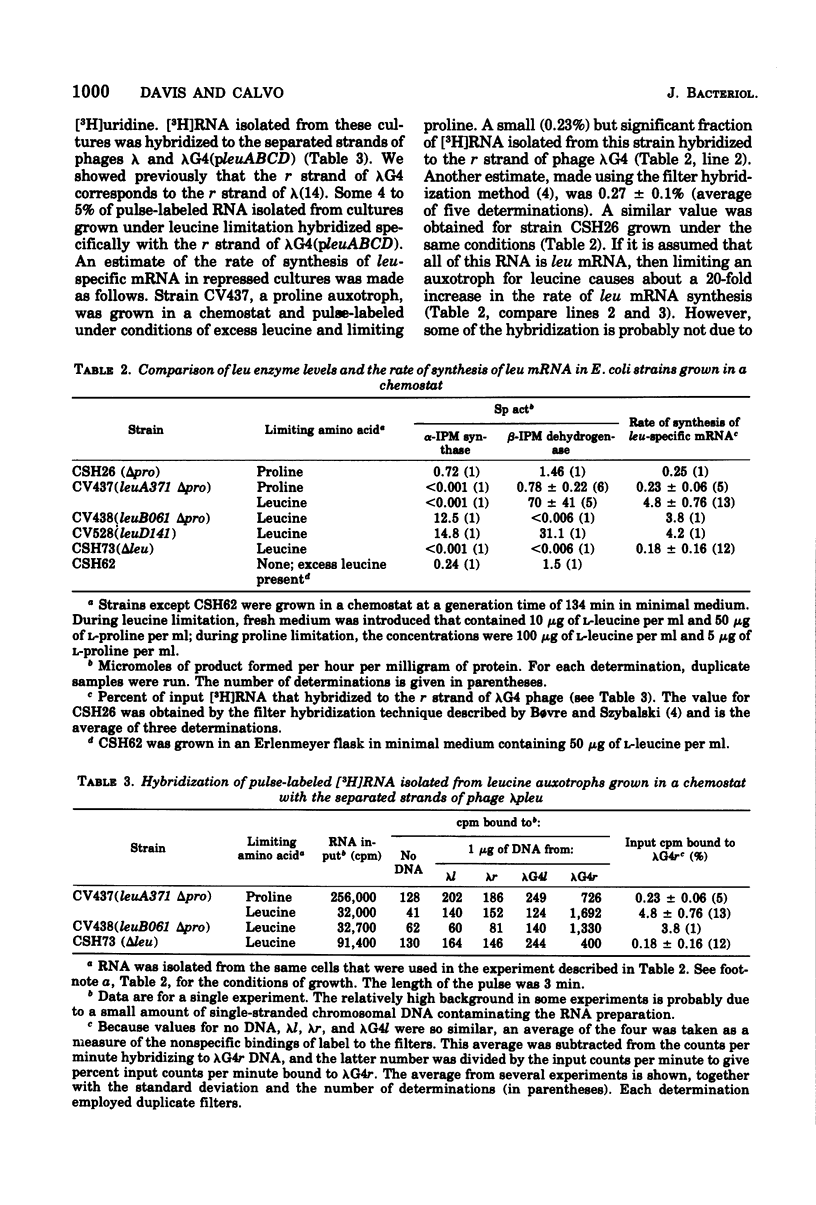

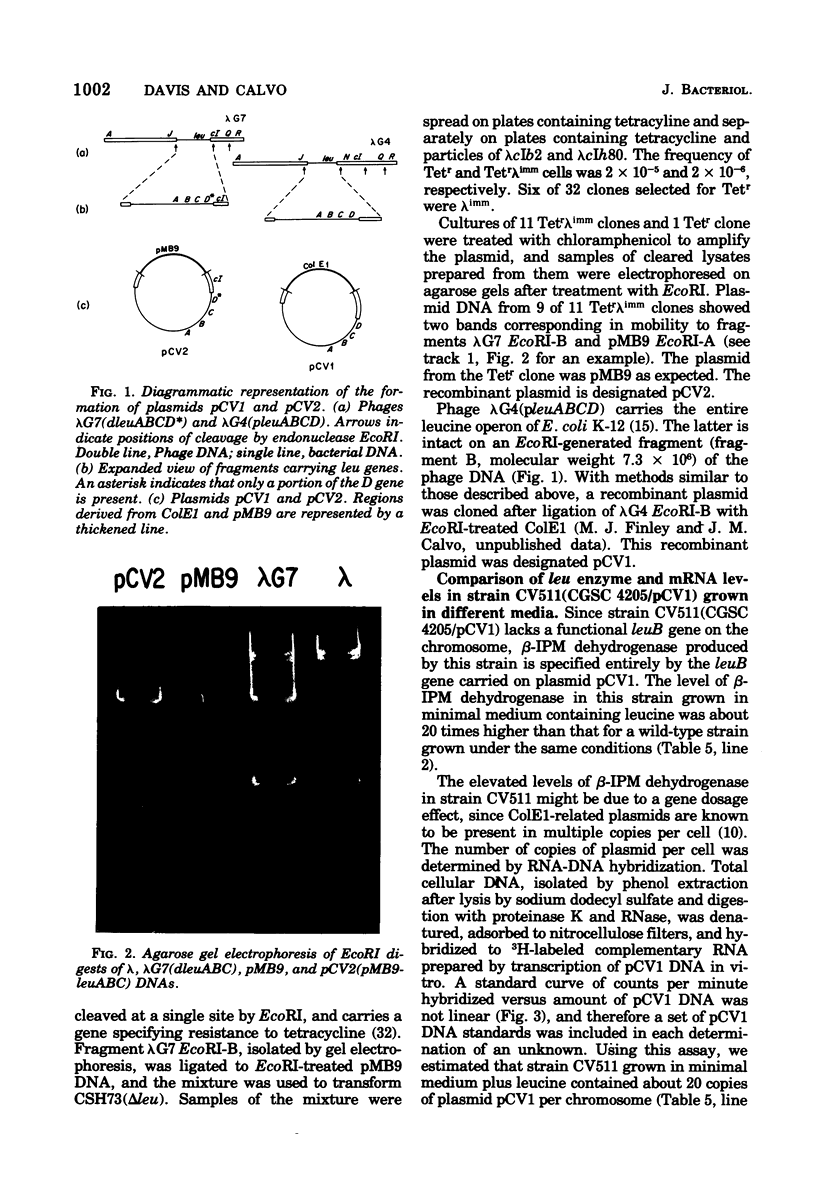
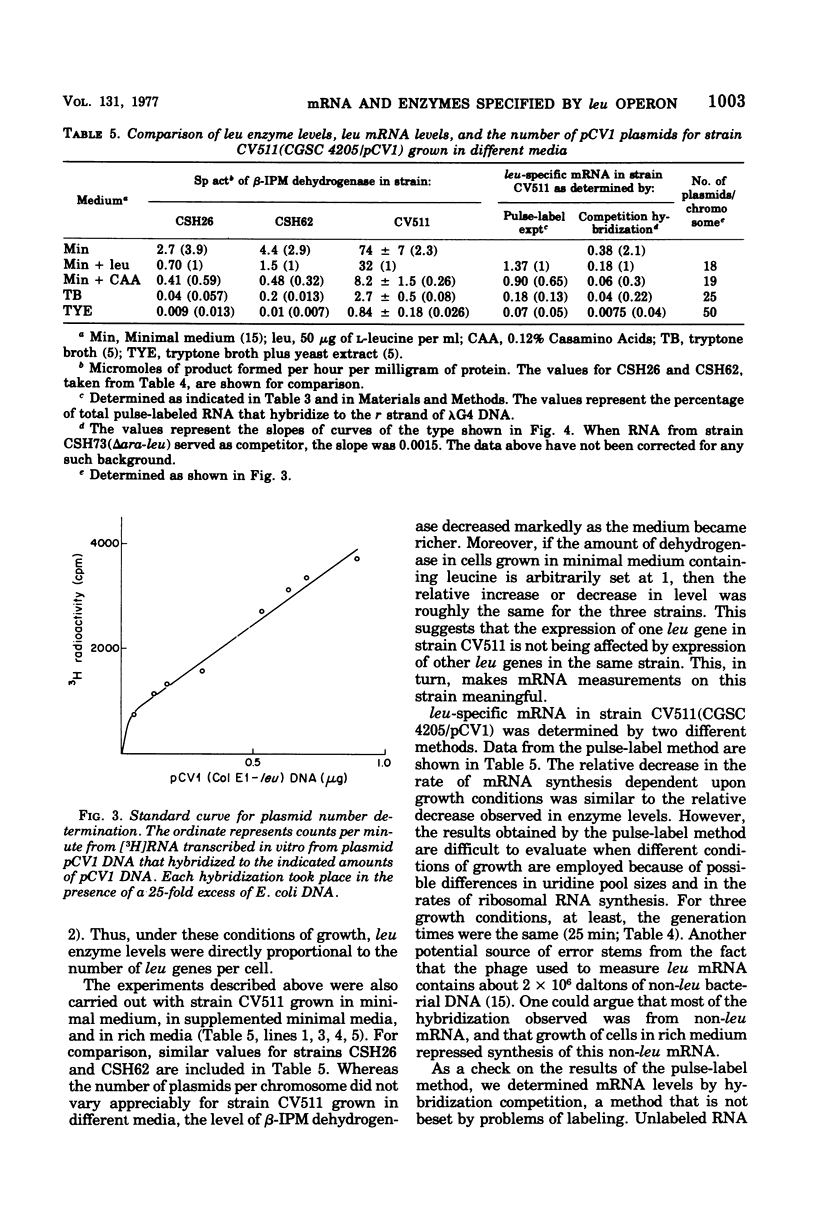



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexander R. R., Calvo J. M., Freundlich M. Mutants of Salmonella typhimurium with an altered leucyl-transfer ribonucleic acid synthetase. J Bacteriol. 1971 Apr;106(1):213–220. doi: 10.1128/jb.106.1.213-220.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artz S. W., Broach J. R. Histidine regulation in Salmonella typhimurium: an activator attenuator model of gene regulation. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3453–3457. doi: 10.1073/pnas.72.9.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand K., Korn L., Lee F., Platt T., Squires C. L., Squires C., Yanofsky C. New features of the regulation of the tryptophan operon. Science. 1975 Jul 4;189(4196):22–26. doi: 10.1126/science.1094538. [DOI] [PubMed] [Google Scholar]
- Brandriss M. C., Calvo J. M. Recognition of an Escherichia operator by a Salmonella repressor. J Bacteriol. 1971 Dec;108(3):1431–1433. doi: 10.1128/jb.108.3.1431-1433.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns R. O., Calvo J., Margolin P., Umbarger H. E. Expression of the leucine operon. J Bacteriol. 1966 Apr;91(4):1570–1576. doi: 10.1128/jb.91.4.1570-1576.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo J. M., Freundlich M., Umbarger H. E. Regulation of branched-chain amino acid biosynthesis in Salmonella typhimurium: isolation of regulatory mutants. J Bacteriol. 1969 Mar;97(3):1272–1282. doi: 10.1128/jb.97.3.1272-1282.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo J. M., Morgolin P., Umbarger H. E. Operator constitutive mutations in the leucine operon of Salmonella typhimurium. Genetics. 1969 Apr;61(4):777–787. doi: 10.1093/genetics/61.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Effect of growth conditions on the formation of the relaxation complex of supercoiled ColE1 deoxyribonucleic acid and protein in Escherichia coli. J Bacteriol. 1972 Jun;110(3):1135–1146. doi: 10.1128/jb.110.3.1135-1146.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese R., Landsberg R., Haar R. A., Umbarger H. E., Ames B. N. Pleiotropy of hisT mutants blocked in pseudouridine synthesis in tRNA: leucine and isoleucine-valine operons. Proc Natl Acad Sci U S A. 1974 May;71(5):1857–1861. doi: 10.1073/pnas.71.5.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress D. E., Kline B. C. Isolation and characterization of Escherichia coli chromosomal mutants affecting plasmid copy number. J Bacteriol. 1976 Feb;125(2):635–642. doi: 10.1128/jb.125.2.635-642.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Court D., Adhya S. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci U S A. 1976 Jun;73(6):1959–1963. doi: 10.1073/pnas.73.6.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. G., Calvo J. M. Isolation and characterization of lambda pleu bacteriophages. J Bacteriol. 1977 Feb;129(2):1078–1090. doi: 10.1128/jb.129.2.1078-1090.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg D., Mikulka T. W., Jones J., Calvo J. M. flrB, a regulatory locus controlling branched-chain amino acid biosynthesis in Salmonella typhimurium. J Bacteriol. 1974 Jun;118(3):942–951. doi: 10.1128/jb.118.3.942-951.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Yanofsky C., Lovett M. A., Helinski D. R. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Dahlberg J. E. Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis. J Biol Chem. 1973 Jul 25;248(14):5024–5032. [PubMed] [Google Scholar]
- Kasai T. Regulation of the expression of the histidine operon in Salmonella typhimurium. Nature. 1974 Jun 7;249(457):523–527. doi: 10.1038/249523a0. [DOI] [PubMed] [Google Scholar]
- Kupersztoch-Portnoy Y. M., Lovett M. A., Helinski D. R. Strand and site specificity of the relaxation event for the relaxation complex of the antibiotic resistance plasmid R6K. Biochemistry. 1974 Dec 31;13(27):5484–5490. doi: 10.1021/bi00724a005. [DOI] [PubMed] [Google Scholar]
- Lavallé R., De Hauwer G. Messenger RNA synthesis during amino acid starvation in Escherichia coli. J Mol Biol. 1968 Oct 28;37(2):269–288. doi: 10.1016/0022-2836(68)90267-2. [DOI] [PubMed] [Google Scholar]
- Lavallé R., De Hauwer G. Tryptophan messenger translation in Escherichia coli. J Mol Biol. 1970 Jul 28;51(2):435–447. doi: 10.1016/0022-2836(70)90153-1. [DOI] [PubMed] [Google Scholar]
- Lavallé R. Regulation at the level of translation in the arginine pathway of Escherichia coli K12. J Mol Biol. 1970 Jul 28;51(2):449–451. doi: 10.1016/0022-2836(70)90154-3. [DOI] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians K. J., Wu R., Stawinski J., Hozumi T., Narang S. A. Cloned synthetic lac operator DNA is biologically active. Nature. 1976 Oct 28;263(5580):744–748. doi: 10.1038/263744a0. [DOI] [PubMed] [Google Scholar]
- Morse D. E., Primakoff P. Relief of polarity in E. coli by "suA". Nature. 1970 Apr 4;226(5240):28–31. doi: 10.1038/226028a0. [DOI] [PubMed] [Google Scholar]
- Neidhardt F. C., Bloch P. L., Smith D. F. Culture medium for enterobacteria. J Bacteriol. 1974 Sep;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzino A. A., Bresalier R. S., Freundlich M. Derepressed levels of the isoleucine-valine and leucine enzymes in his T 1504, a strain of Salmonella typhimurium with altered leucine transfer ribonucleic acid. J Bacteriol. 1974 Feb;117(2):449–455. doi: 10.1128/jb.117.2.449-455.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose J. K., Squires C. L., Yanofsky C., Yang H. L., Zubay G. Regulation of in vitro transcription of the tryptophan operon by purified RNA polymerase in the presence of partially purified repressor and tryptophan. Nat New Biol. 1973 Oct 3;245(144):133–137. doi: 10.1038/newbio245133a0. [DOI] [PubMed] [Google Scholar]
- Somers J. M., Amzallag A., Middleton R. B. Genetic fine structure of the leucine operon of Escherichia coli K-12. J Bacteriol. 1973 Mar;113(3):1268–1272. doi: 10.1128/jb.113.3.1268-1272.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth J. J., Umbarger H. E. Effect of isoleucine, valine, or leucine starvation on the potential for formation of the branched-chain amino acid biosynthetic enzymes. J Bacteriol. 1973 Nov;116(2):548–561. doi: 10.1128/jb.116.2.548-561.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H. L., Kessler D. P. Genetic analysis of the leucine region in Escherichia coli B-r: gene-enzyme assignments. J Bacteriol. 1974 Jan;117(1):63–72. doi: 10.1128/jb.117.1.63-72.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gussin G. N. Genetic characterization of a prm- mutant of bacteriophage lambda. Virology. 1973 Nov;56(1):300–312. doi: 10.1016/0042-6822(73)90308-5. [DOI] [PubMed] [Google Scholar]