Abstract
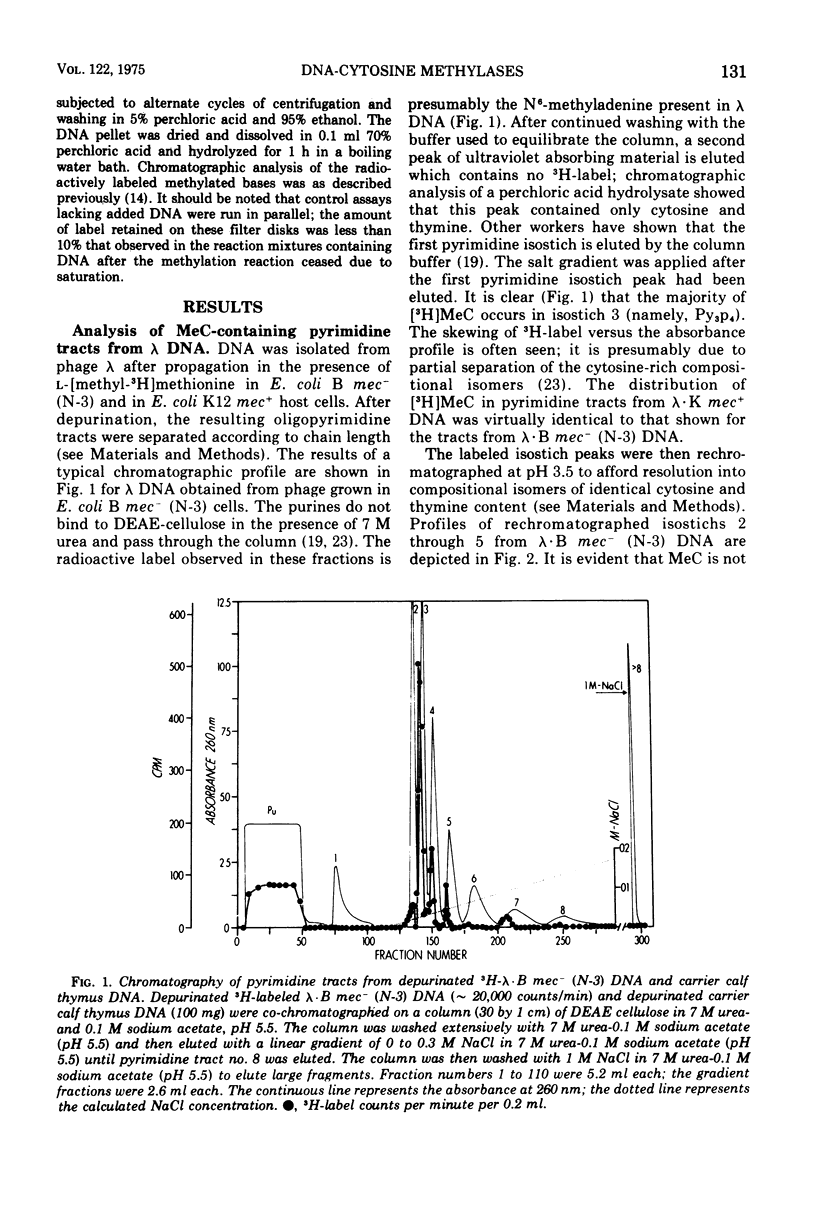
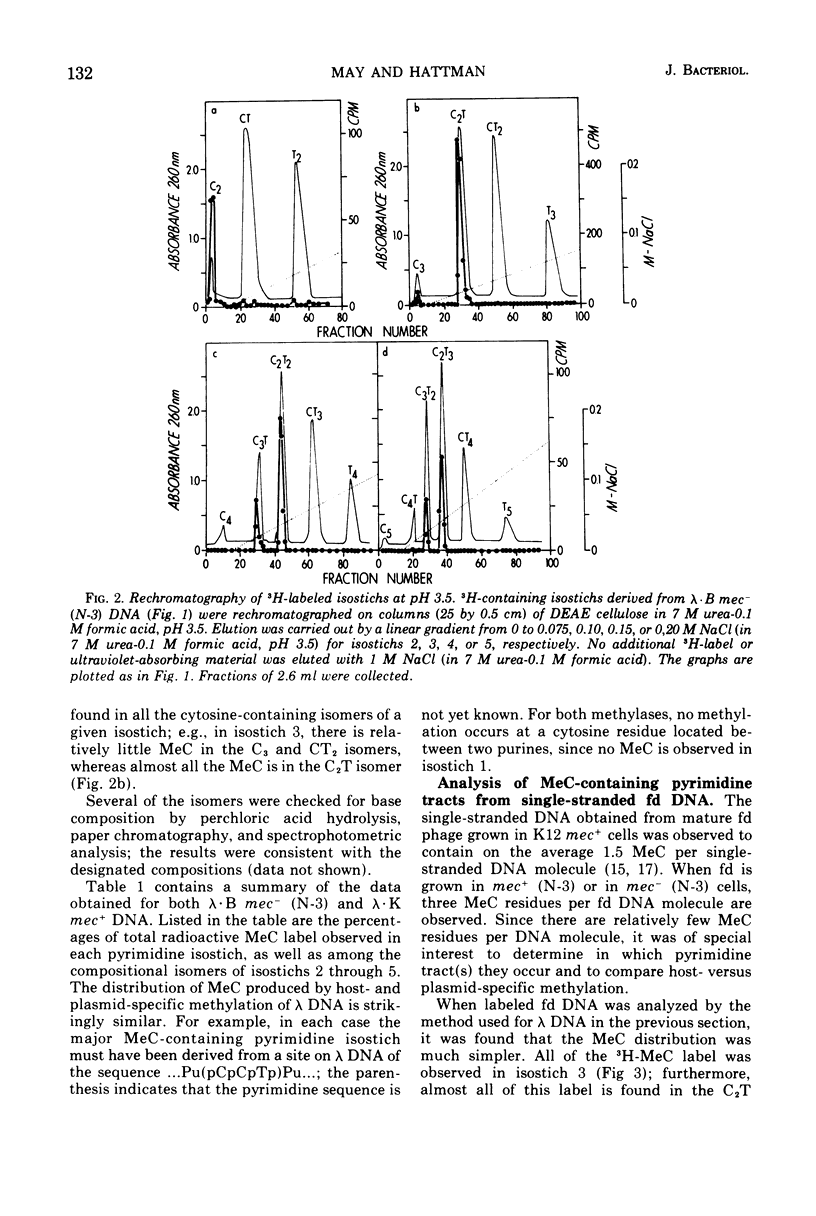
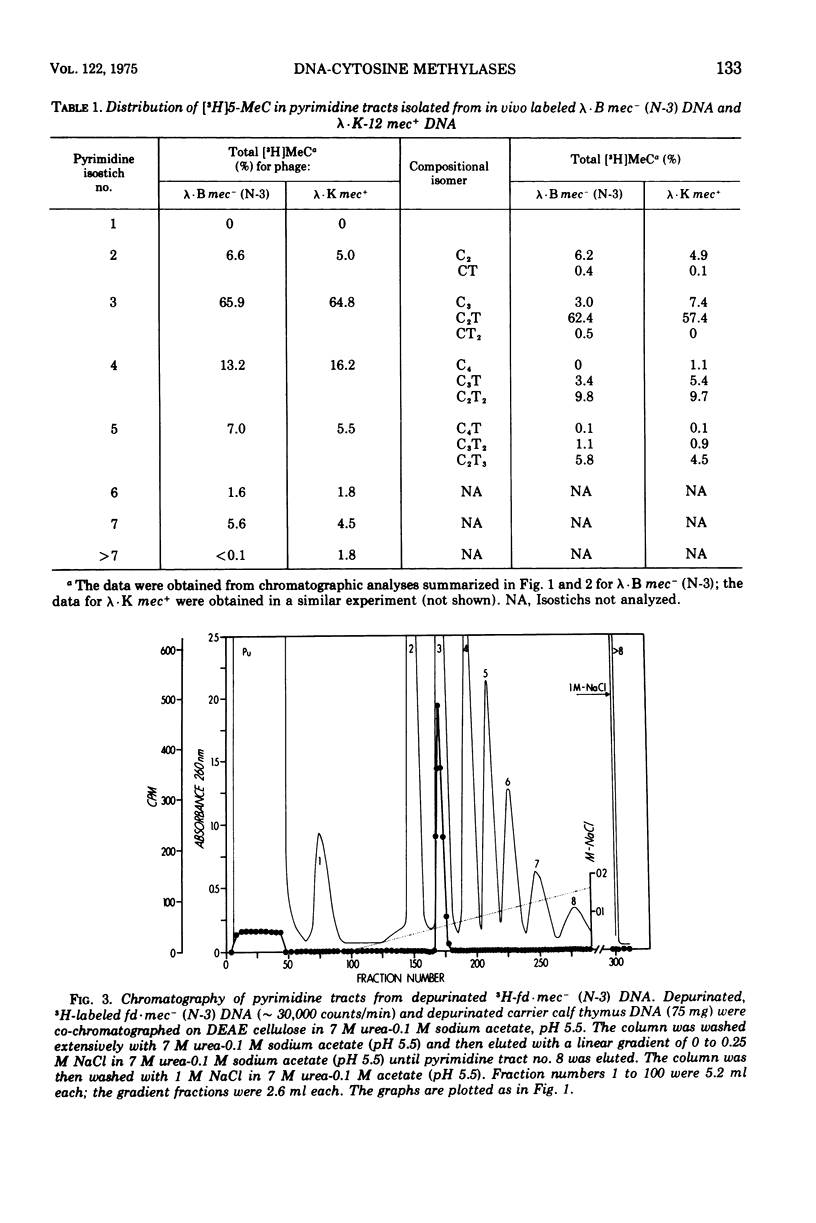
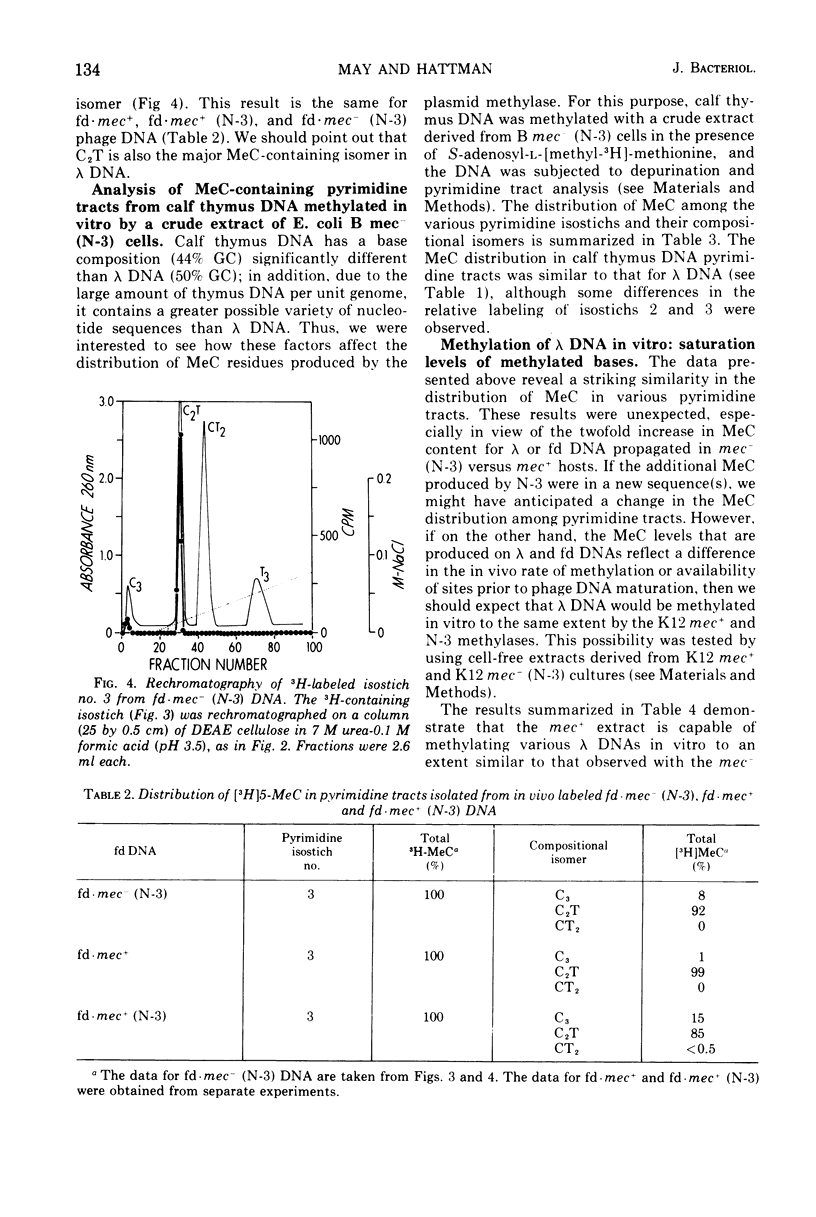
Deoxyribonucleic acid (DNA)-cytosine methylation specified by the wild-type Escherichia coli K 12 mec+ gene and by the N-3 drug resistance (R) factor was studied in vivo and in vitro. Phage lambda and fd were propagated in the presence of L-[methyl-3H]methionine in various host bacteria. The in vivo labeled DNA was isolated from purified phage and depurinated by formic acid-diphenylamine treatment. The resulting pyrimidine oligonucleotide tracts were separated according to size and base composition by chromatography on diethylaminoethyl-cellulose in 7 M urea at pH 5.5 and 3.5, respectively. The distribution of labeled 5-methylcytosine in DNA pyrimidine tracts was identical for phage grown in mec+ and mec minus (N-3) cells. For phage lambda the major 5-methylcytosine containing tract was the tripyrimidine, C2T; for both fd-mec minus (N-3) DNA and fd-mec+DNA, C2T was the sole 5-methylcytosine-containing tract. When various lambda DNAs were methylated to saturation in vitro by crude extracts from mec+ and mec minus (N-3) cells, the extent of cytosine methylation was the same. This is in contrast to in vivo methylation where lambda-mec minus (N-3) DNA contains twice as many 5-methylcytosines per genome as lambda-mec+ DNA. Therefore, we suggest that the K12 met+ cytosine methylase and the N-3 plasmid modification methylase are capable of recognizing the same nucleotide sequences, but that the in vivo methylation rate is lower in mec+ cells.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arber W. Host specificity of DNA produced by Escherichia coli. 9. Host-controlled modification of bacteriophage fd. J Mol Biol. 1966 Oct;20(3):483–496. doi: 10.1016/0022-2836(66)90004-0. [DOI] [PubMed] [Google Scholar]
- Arber W., Linn S. DNA modification and restriction. Annu Rev Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- BURTON K., PETERSEN G. B. The frequencies of certain sequences of nucleotides in deoxyribonucleic acid. Biochem J. 1960 Apr;75:17–27. doi: 10.1042/bj0750017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister D., Glover S. W. Restriction and modification of bacteriophages by R+ strains of Escherichia coli K12. Biochem Biophys Res Commun. 1968 Mar 27;30(6):735–738. doi: 10.1016/0006-291x(68)90575-5. [DOI] [PubMed] [Google Scholar]
- Bannister D., Glover S. W. The isolation and properties of non-restricting mutants of two different host specificities associated with drug resistance factors. J Gen Microbiol. 1970 Apr;61(1):63–71. doi: 10.1099/00221287-61-1-63. [DOI] [PubMed] [Google Scholar]
- Bickle T., Arber W. Host-controlled restriction and modification of filamentous 1- and F-specific bacteriophages. Virology. 1969 Nov;39(3):605–607. doi: 10.1016/0042-6822(69)90112-3. [DOI] [PubMed] [Google Scholar]
- Bigger C. H., Murray K., Murray N. E. Recognition sequence of a restriction enzyme. Nat New Biol. 1973 Jul 4;244(131):7–10. doi: 10.1038/newbio244007a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Chow L. T., Dugaiczyk A., Hedgpeth J., Goodman H. M. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat New Biol. 1973 Jul 11;244(132):40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- Boyer H. W. DNA restriction and modification mechanisms in bacteria. Annu Rev Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- DOSKOCIL J., SORMO'VA Z. THE OCCURRENCE OF 5-METHYLCYTOSINE IN BACTERIAL DEOXYRIBONUCLEIC ACIDS. Biochim Biophys Acta. 1965 Mar 15;95:513–515. [PubMed] [Google Scholar]
- Doskocil J., Sormová Z. The sequences of 5-methylcytosine in the DNA of Escherichia coli. Biochem Biophys Res Commun. 1965 Jul 26;20(3):334–339. doi: 10.1016/0006-291x(65)90369-4. [DOI] [PubMed] [Google Scholar]
- Fujimoto D., Srinivasan P. R., Borek E. On the nature of the deoxyribonucleic acid methylases. Biological evidence for the multiple nature of the enzymes. Biochemistry. 1965 Dec;4(12):2849–2855. doi: 10.1021/bi00888a041. [DOI] [PubMed] [Google Scholar]
- Gough M., Lederberg S. Methylated bases in the host-modified deoxyribonucleic acid of Escherichia coli and bacteriophage lambda. J Bacteriol. 1966 Apr;91(4):1460–1468. doi: 10.1128/jb.91.4.1460-1468.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Gold E., Plotnik A. Methylation of cytosine residues in DNA controlled by a drug resistance factor (host-induced modification-R factors-N 6 -methyladenine-5-methylcytosine). Proc Natl Acad Sci U S A. 1972 Jan;69(1):187–190. doi: 10.1073/pnas.69.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Plasmid-controlled variation in the content of methylated bases in bacteriophage lambda deoxyribonucleic acid. J Virol. 1972 Sep;10(3):356–361. doi: 10.1128/jvi.10.3.356-361.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S. Plasmid-controlled variation in the content of methylated bases in single-stranded DNA bacteriophages M13 and fd. J Mol Biol. 1973 Mar 15;74(4):749–752. doi: 10.1016/0022-2836(73)90064-8. [DOI] [PubMed] [Google Scholar]
- Hattman S., Schlagman S., Cousens L. Isolation of a mutant of Escherichia coli defective in cytosine-specific deoxyribonucleic acid methylase activity and in partial protection of bacteriophage lambda against restriction by cells containing the N-3 drug-resistance factor. J Bacteriol. 1973 Sep;115(3):1103–1107. doi: 10.1128/jb.115.3.1103-1107.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo C., Nash H. A. Methylation pattern of lambda deoxyribonucleic acid. J Virol. 1972 Nov;10(5):937–942. doi: 10.1128/jvi.10.5.937-942.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer P. E., Saunders G. F. Distribution of pyrimidine sequences in bacteriophage TP-84 deoxyribonucleic acid. Biochemistry. 1972 Apr 25;11(9):1562–1568. [PubMed] [Google Scholar]
- LEDINKO N. OCCURRENCE OF 5-METHYLDEOXYCYTIDYLATE IN THE DNA OF PHAGE LAMBDA. J Mol Biol. 1964 Sep;9:834–835. doi: 10.1016/s0022-2836(64)80191-1. [DOI] [PubMed] [Google Scholar]
- Lederberg S. 5-Methylcytosine in the host-modified DNA of Escherichia coli and phage lambda. J Mol Biol. 1966 May;17(1):293–297. doi: 10.1016/s0022-2836(66)80111-0. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushizky G. W., Sober H. A. Chromatography of tri- and tetranucleotides from pancreatic ribonuclease digests of ribonucleic acid. Biochem Biophys Res Commun. 1964;14:276–279. doi: 10.1016/0006-291x(64)90449-8. [DOI] [PubMed] [Google Scholar]
- Schlagman S., Hattman S. Mutants of the N-3 R-factor conditionally defective in hspII modification and deoxyribonucleic acid-cytosine methylase activity. J Bacteriol. 1974 Oct;120(1):234–239. doi: 10.1128/jb.120.1.234-239.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Watanabe T., Fukasawa T. Mechanism of host-controlled restriction of bacteriophage lambda by R factors in Escherichia coli K12. Virology. 1968 Feb;34(2):290–302. doi: 10.1016/0042-6822(68)90239-0. [DOI] [PubMed] [Google Scholar]
- Yoshimori R., Roulland-Dussoix D., Boyer H. W. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J Bacteriol. 1972 Dec;112(3):1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]


