Abstract
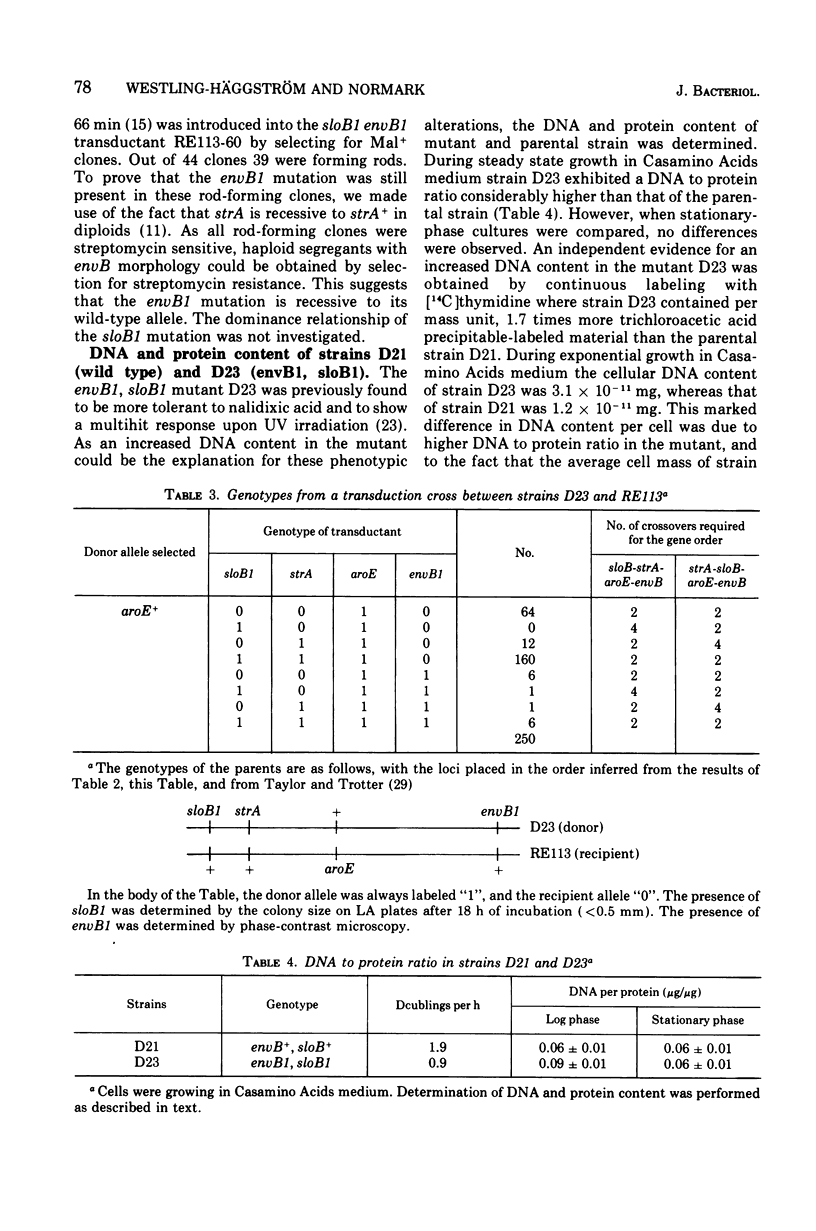
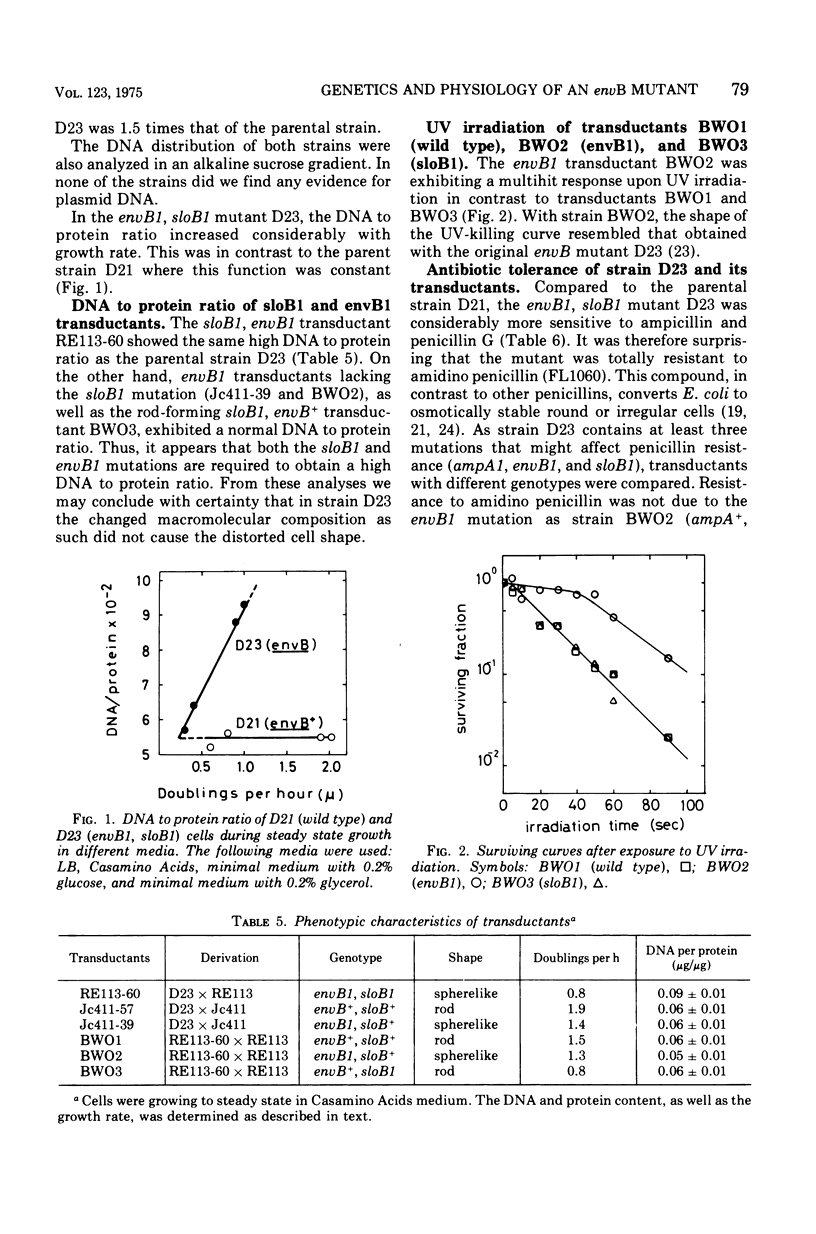
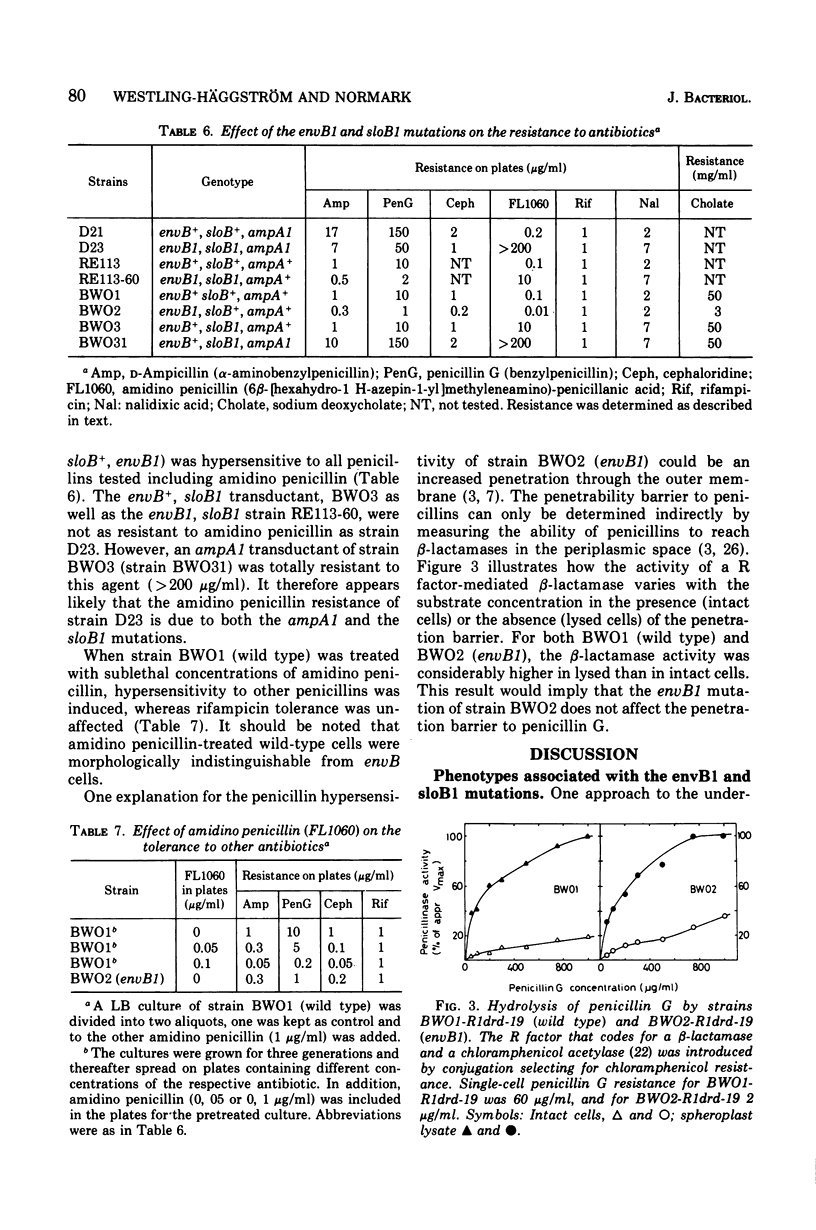
The envB1 mutation mediating a distorted cell morphology of Escherichia coliK-12 was cotransducible with strA, aroE, aspB, and argG. The mapping data is consistent with a gene location for envB around 62.5 min. In partial diploids envB1 was recessive to its wild-type allele. The original envB mutant contained a second mutation in a locus denoted sloB close to strA. The following gene order is suggested: sloB-strA-aroE-envB-aspB-argG. The sloB1 mutation caused a marked reduction in the growth rate of both envB and envB+ strains. Moreover, this mutation in the presence of envB1 appears to increase the ratio between deoxyribonucleic acid and protein in cells growing in rich medium. The phenotypic properties of envB1, sloB+ and envB+ transductants were characterized. Cells with envB1, sloB+ genotype were hypersensitive to several penicillins including the beta-lactam compound, amidino penicillin. Penicillin hypersensitivity could not be explained by increased outer membrane penetrability. The original envB mutant (envB1,SLOB1), as well as envB1, sloB1 or envB+, SLOB1 transductants were resistant to amidino penicillin. Resistance was explained by the slow growth rate mediated by the sloB1 mutation. The similarity between envB cells and wild-type cells treated with sublethal concentrations of amidino penicillin was emphasized.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom G. D., Gumpert J., Normark S., Schuhmann E., Taubeneck U., Westling B. Morphology and growth pattern of a rod-negative envB mutant of Escherichia coli K12. Z Allg Mikrobiol. 1974;14(6):465–477. doi: 10.1002/jobm.3630140603. [DOI] [PubMed] [Google Scholar]
- Boman H. G., Nordström K., Normark S. Penicillin resistance in Escherichia coli K12: synergism between penicillinases and a barrier in the outer part of the envelope. Ann N Y Acad Sci. 1974 May 10;235(0):569–586. doi: 10.1111/j.1749-6632.1974.tb43291.x. [DOI] [PubMed] [Google Scholar]
- Cerdá-Olmedo E., Hanawalt P. C. The replication of the Escherichia coli chromosome studied by sequential nitrosoguanidine mutagenesis. Cold Spring Harb Symp Quant Biol. 1968;33:599–607. doi: 10.1101/sqb.1968.033.01.066. [DOI] [PubMed] [Google Scholar]
- Dennis P. P., Bremer H. Macromolecular composition during steady-state growth of Escherichia coli B-r. J Bacteriol. 1974 Jul;119(1):270–281. doi: 10.1128/jb.119.1.270-281.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson-Grennberg K. G. Resistance of Escherichia coli to penicillins. II. An improved mapping of the ampA gene. Genet Res. 1968 Oct;12(2):147–156. doi: 10.1017/s0016672300011769. [DOI] [PubMed] [Google Scholar]
- Henning U., Höhn B., Sonntag I. Cell envelope and shape of Escherichia coli K12. The ghost membrane. Eur J Biochem. 1973 Nov 1;39(1):27–36. doi: 10.1111/j.1432-1033.1973.tb03099.x. [DOI] [PubMed] [Google Scholar]
- Henning U., Rehn K., Braun V., Höhn B. Cell envelope and shape of Escherichia coli K12. Properties of a temperature-sensitive rod mutant. Eur J Biochem. 1972 Apr 24;26(4):570–586. doi: 10.1111/j.1432-1033.1972.tb01800.x. [DOI] [PubMed] [Google Scholar]
- Kadner R. J., Maas W. K. Regulatory gene mutations affecting arginine biosynthesis in Escherichia coli. Mol Gen Genet. 1971;111(1):1–14. doi: 10.1007/BF00286549. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lazdunski C., Shapiro B. M. Isolation and some properties of cell envelope altered mutants of Escherichia coli. J Bacteriol. 1972 Aug;111(2):495–498. doi: 10.1128/jb.111.2.495-498.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdunski C., Shaprio B. M. Relationship between permeability, cell division, and murein metabolism in a mutant of Escherichia coli. J Bacteriol. 1972 Aug;111(2):499–509. doi: 10.1128/jb.111.2.499-509.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindström E. B., Nordström K. Automated method for determination of penicillins, cephalosporins, and penicillinases. Antimicrob Agents Chemother. 1972 Feb;1(2):100–106. doi: 10.1128/aac.1.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund F., Tybring L. 6 -amidinopenicillanic acids--a new group of antibiotics. Nat New Biol. 1972 Apr 5;236(66):135–137. doi: 10.1038/newbio236135a0. [DOI] [PubMed] [Google Scholar]
- Matsuhashi S., Kamiryo T., Blumberg P. M., Linnett P., Willoughby E., Strominger J. L. Mechanism of action and development of resistance to a new amidino penicillin. J Bacteriol. 1974 Feb;117(2):578–587. doi: 10.1128/jb.117.2.578-587.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa H., Hayakawa K., Sato T., Imahori K. Characterization and genetic analysis of a mutant of Escherichia coli K-12 with rounded morphology. J Bacteriol. 1973 Jul;115(1):436–442. doi: 10.1128/jb.115.1.436-442.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior N. H., Blom J., Tybring L., Birch-Andersen A. Light and electron microscopy of the early response of Escherichia coli to a 6beta-amidinopenicillanic acid (FL 1060). Acta Pathol Microbiol Scand B Microbiol Immunol. 1973 Aug;81(4):393–407. doi: 10.1111/j.1699-0463.1973.tb02222.x. [DOI] [PubMed] [Google Scholar]
- Meynell E., Datta N. Mutant drug resistant factors of high transmissibility. Nature. 1967 May 27;214(5091):885–887. doi: 10.1038/214885a0. [DOI] [PubMed] [Google Scholar]
- Normark S. Mutation in Escherichia coli K-12 mediating spherelike envelopes and changes tolerance to ultraviolet irradiation and some antibiotics. J Bacteriol. 1969 Jun;98(3):1274–1277. doi: 10.1128/jb.98.3.1274-1277.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. T., Burman L. FL-1060: a new penicillin with a unique mode of action. Biochem Biophys Res Commun. 1973 Apr 16;51(4):863–868. doi: 10.1016/0006-291x(73)90006-5. [DOI] [PubMed] [Google Scholar]
- Reiner A. M. Isolation and mapping of polynucleotide phosphorylase mutants of Escherichia coli. J Bacteriol. 1969 Mar;97(3):1431–1436. doi: 10.1128/jb.97.3.1431-1436.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond M. H., Curtis N. A. The interplay of beta-lactamases and intrinsic factors in the resistance of gram-negative bacteria to penicillins and cephalosporins. Ann N Y Acad Sci. 1974 May 10;235(0):553–568. doi: 10.1111/j.1749-6632.1974.tb43290.x. [DOI] [PubMed] [Google Scholar]
- Silver R. P., Falkow S. Specific labeling and physical characterization of R-factor deoxyribonucleic acid in Escherichia coli. J Bacteriol. 1970 Oct;104(1):331–339. doi: 10.1128/jb.104.1.331-339.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]


