Abstract
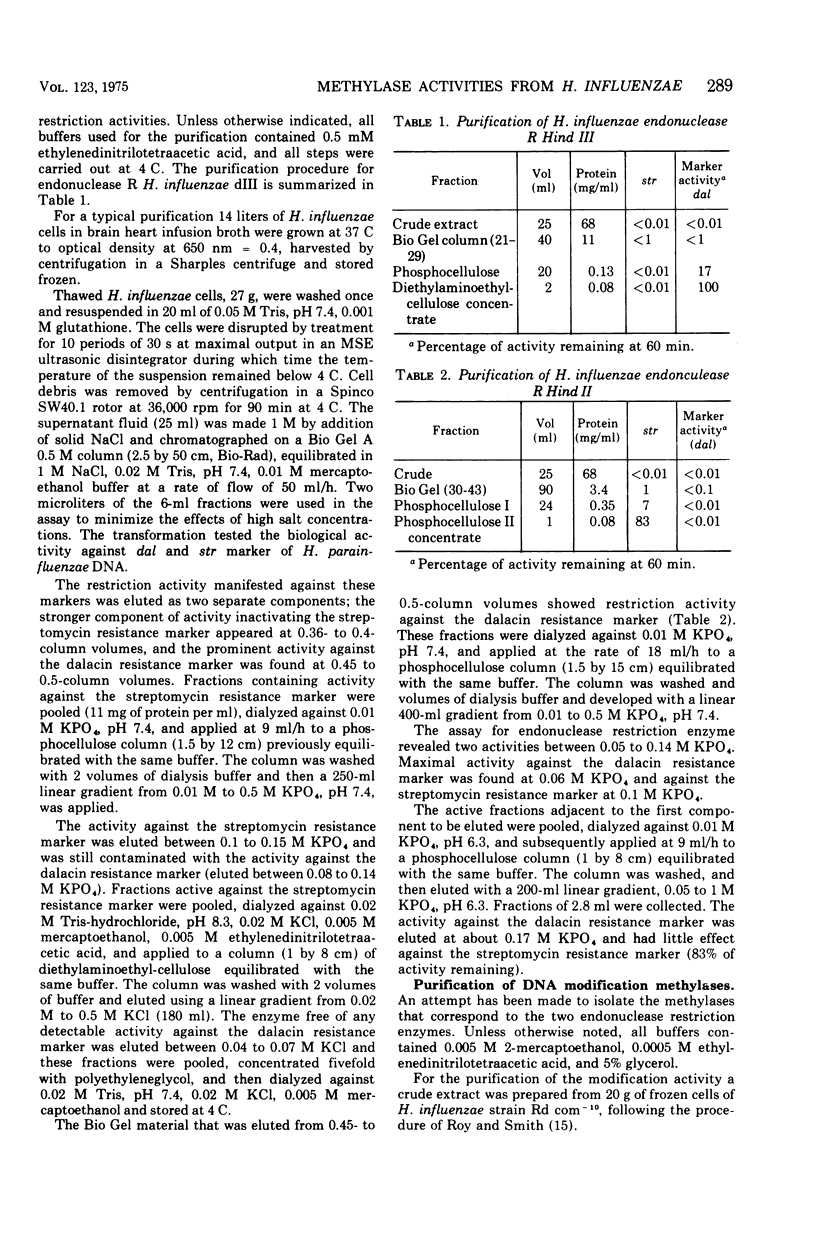
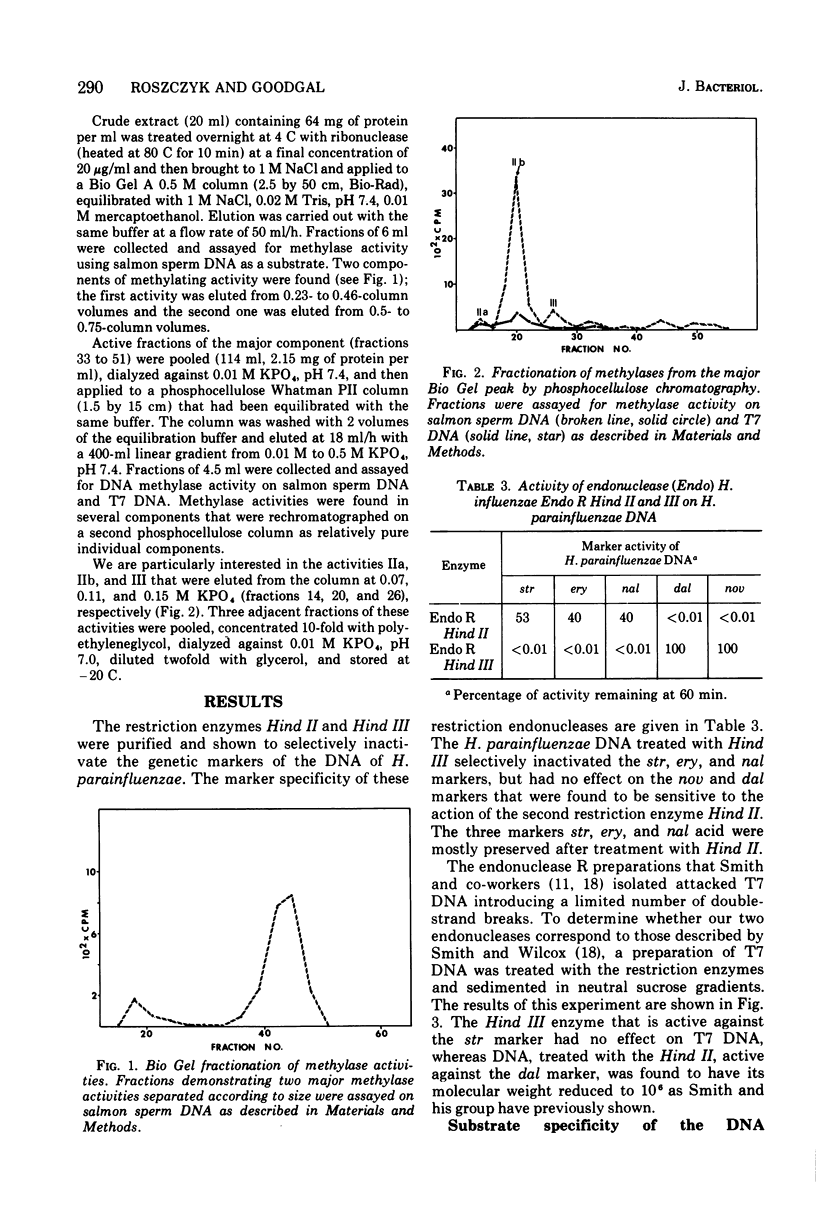
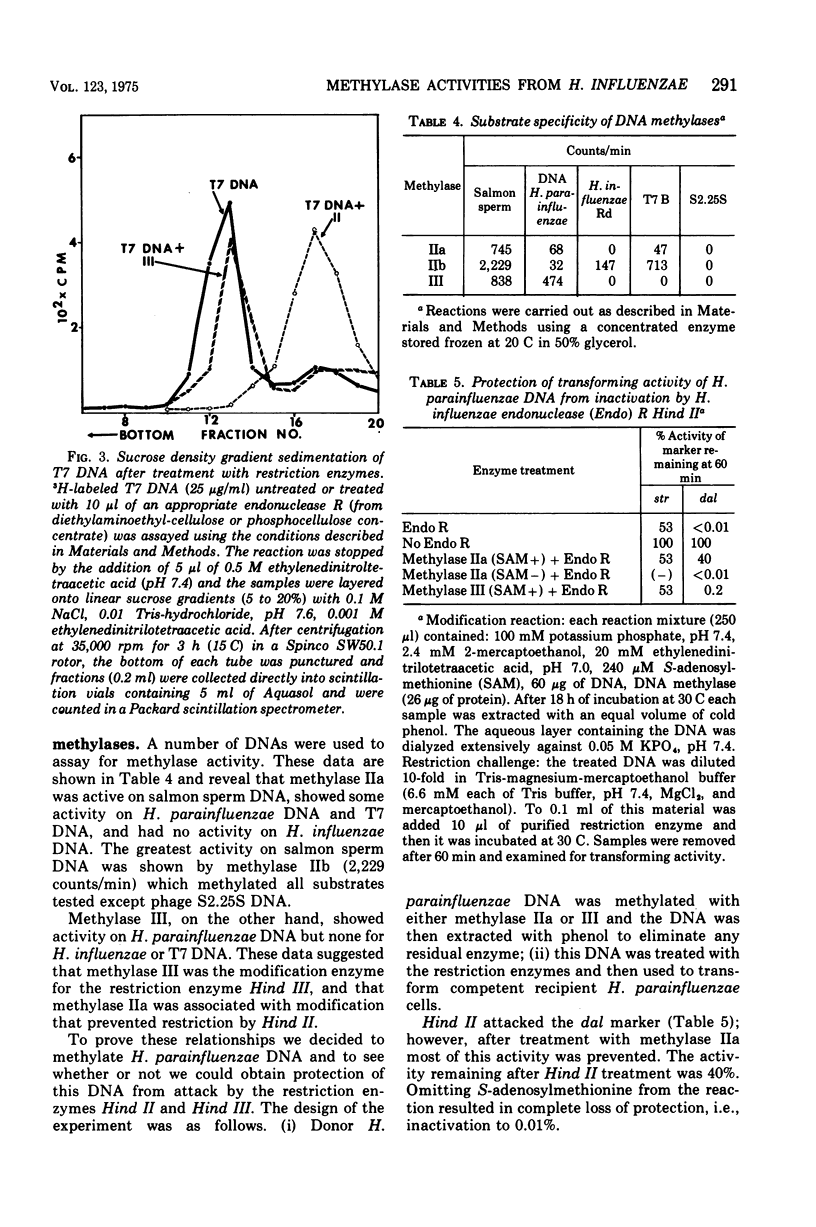
Specific methylases that have the properties of deoxyribonucleic acid (DNA) modification enzymes have been isolated from Haemophilus influenzae strain Rd. Two activities ((Methylase IIa and methylase III) were found to protect transforming DNA of H. parainfluenzae from the action of H. influenzae restriction enzymes. To determine the specificty of the protection, a procedure based on biological activity was developed for the separation and purification of the restriction endonucleases from H. influenzae strain Rd. Two endonuclease R activities presumably corresponding to Hind II and Hind III (P. H. Roy and H. O. Smith, 1973; H. O. Smith and K. W. Wilcox, 1970) were characterized by differences in their chromatographic properties, ability to attack T7 DNA, and inactivation of the transforming activity of different markers of H. parainfluenzae DNA. One endonuclease R enzyme (Hind II) attacked T7 DNA and was found to inactivate the dalacin resistance marker (smaller than 0.01% activity remaining) with only a slight effect on the streptomycin resistance marker (83% activity remaining). Methylase IIa treatment protected 40% of the dalacin resistance marker of H. parainfluenzae DNA from inactivation by Hind II. The other restriction activity (Hind III) was inert towards T7 DNA and inactivated the streptomycin resistance marker of H. parainfluenzae DNA (smaller than 0.01% activity remaining) without any effect on the dalacin resistance marker. The methylation of H. parainfluenzae DNA accomplished by methylase III protected 60% of the transforming activity of the streptomycin resistance marker of H. parainfluenzae DNA from the action of Hind III.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARBER W., DUSSOIX D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J Mol Biol. 1962 Jul;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- DUSSOIX D., ARBER W. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J Mol Biol. 1962 Jul;5:37–49. doi: 10.1016/s0022-2836(62)80059-x. [DOI] [PubMed] [Google Scholar]
- GOLD M., HURWITZ J., ANDERS M. The enzymatic methylation of RNA and DNA. Biochem Biophys Res Commun. 1963 Apr 23;11:107–114. doi: 10.1016/0006-291x(63)90075-5. [DOI] [PubMed] [Google Scholar]
- GOODGAL S. H., HERRIOTT R. M. Studies on transformations of Hemophilus influenzae. I. Competence. J Gen Physiol. 1961 Jul;44:1201–1227. doi: 10.1085/jgp.44.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover S. W., Piekarowicz A. Host specificity of DNA in Haemophilus influenzae: restriction and modification in strain Rd. Biochem Biophys Res Commun. 1972 Feb 25;46(4):1610–1617. doi: 10.1016/0006-291x(72)90793-0. [DOI] [PubMed] [Google Scholar]
- Gold M., Hurwitz J., Anders M. THE ENZYMATIC METHYLATION OF RNA AND DNA, II. ON THE SPECIES SPECIFICITY OF THE METHYLATION ENZYMES. Proc Natl Acad Sci U S A. 1963 Jul;50(1):164–169. doi: 10.1073/pnas.50.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther J. K., Goodgal S. H. An exonuclease specific for double stranded deoxyribonucleic acid. J Biol Chem. 1970 Oct 25;245(20):5341–5349. [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. M., Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOZINSKI A. W., SZYBALSKI W. Dispersive transfer of the parental DNA molecule to the progeny of phage phiX-174. Virology. 1959 Oct;9:260–274. doi: 10.1016/0042-6822(59)90119-9. [DOI] [PubMed] [Google Scholar]
- Kelly T. J., Jr, Smith H. O. A restriction enzyme from Hemophilus influenzae. II. J Mol Biol. 1970 Jul 28;51(2):393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- NICKEL L., GOODGAL S. H. EFFECT OF INTERSPECIFIC TRANSFORMATION ON LINKAGE RELATIONSHIPS OF MARKERS IN HAEMOPHILUS INFLUENZAE AND HAEMOPHILUS PARAINFLUENZAE. J Bacteriol. 1964 Dec;88:1538–1544. doi: 10.1128/jb.88.6.1538-1544.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. I. Purification and properties. J Mol Biol. 1973 Dec 25;81(4):427–444. doi: 10.1016/0022-2836(73)90515-9. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. II. Partial recognition site base sequences. J Mol Biol. 1973 Dec 25;81(4):445–459. doi: 10.1016/0022-2836(73)90516-0. [DOI] [PubMed] [Google Scholar]
- Shapiro S. K., Ehninger D. J. Methods for the analysis and preparation of adenosylmethionine and adenosylhomocysteine. Anal Biochem. 1966 May;15(2):323–333. doi: 10.1016/0003-2697(66)90038-8. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Wilcox K. W. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J Mol Biol. 1970 Jul 28;51(2):379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- Smith J. D., Arber W., Kühnlein U. Host specificity of DNA produced by Escherichia coli. XIV. The role of nucleotide methylation in in vivo B-specific modification. J Mol Biol. 1972 Jan 14;63(1):1–8. doi: 10.1016/0022-2836(72)90517-7. [DOI] [PubMed] [Google Scholar]
- Yuan R., Meselson M. A specific complex between a restriction endonuclease and its DNA substrate. Proc Natl Acad Sci U S A. 1970 Feb;65(2):357–362. doi: 10.1073/pnas.65.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]


