Abstract
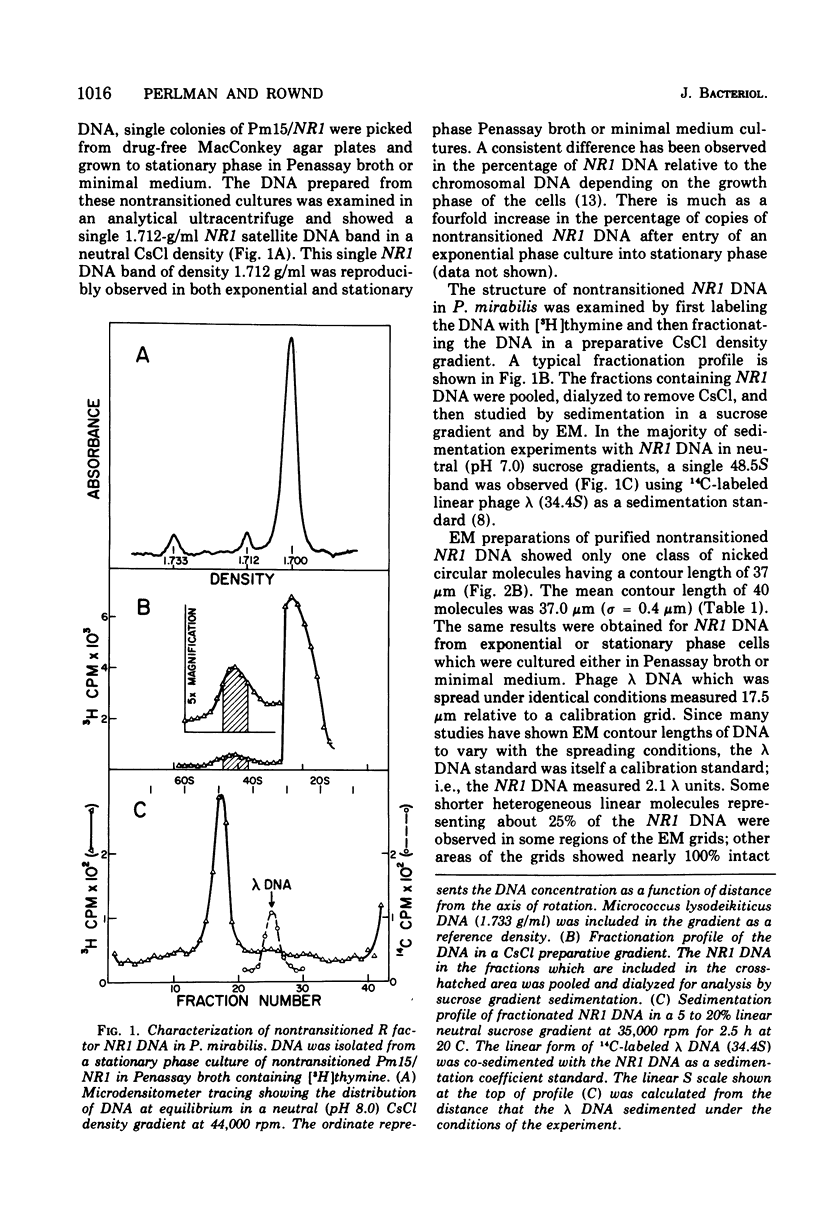
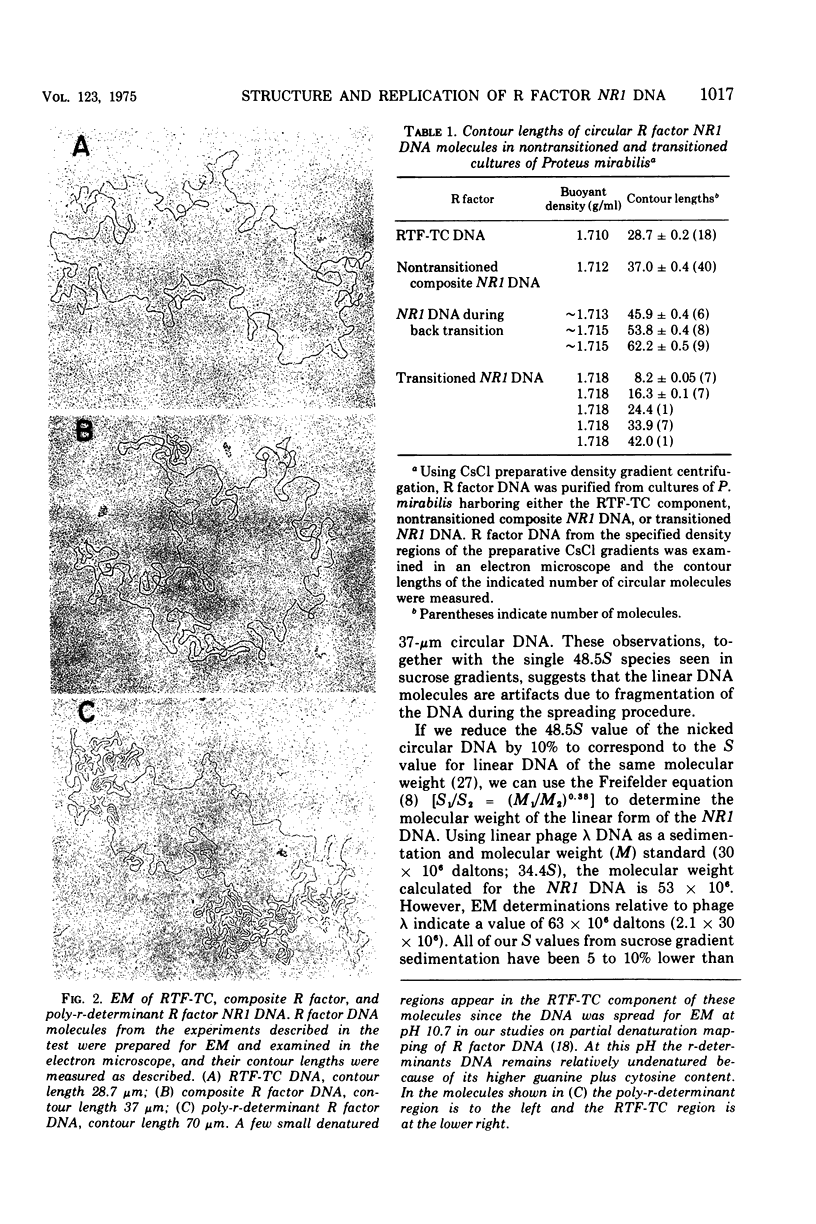
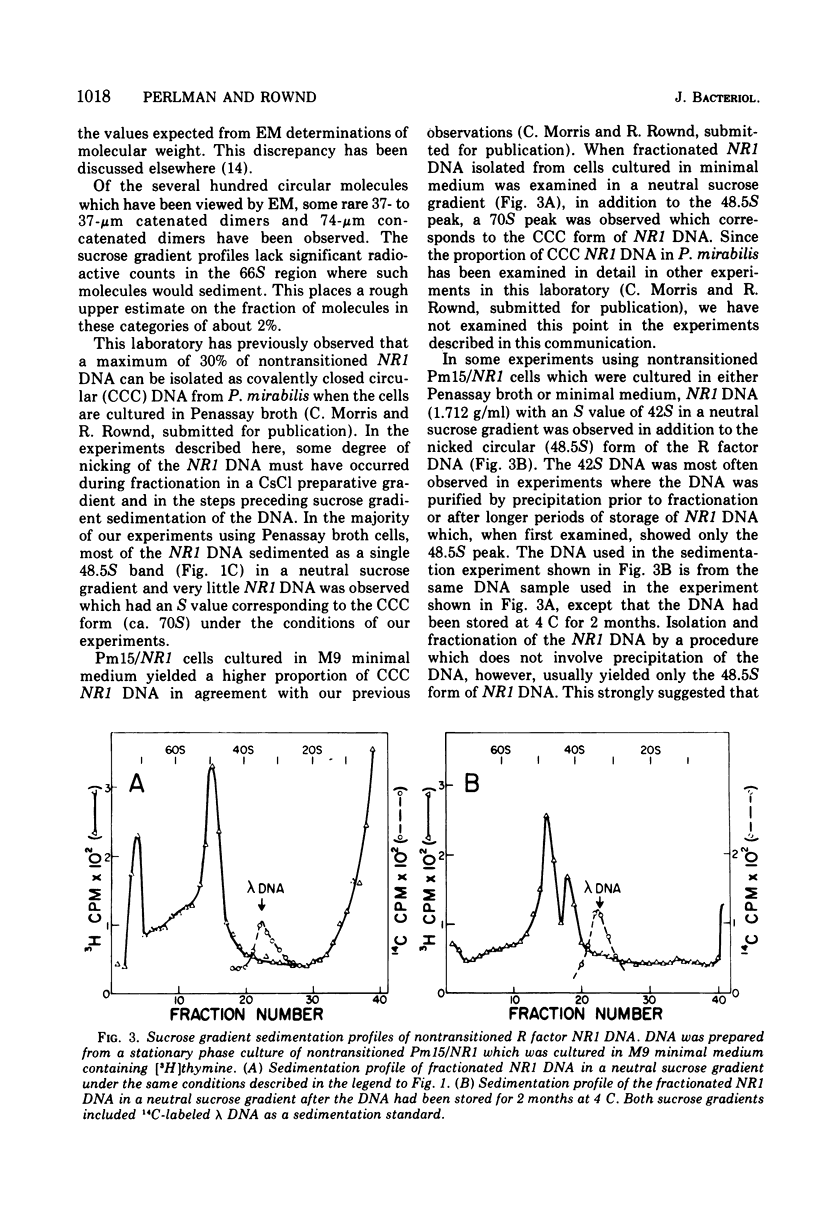
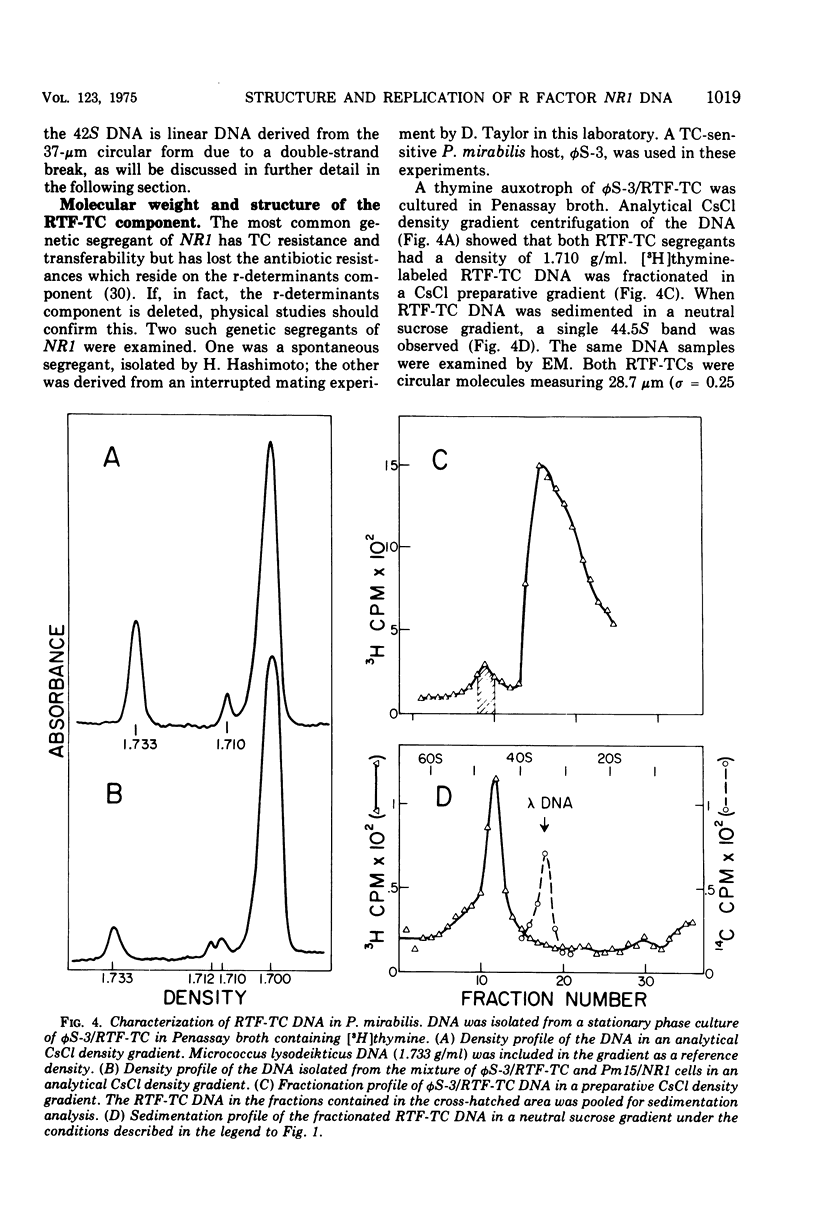
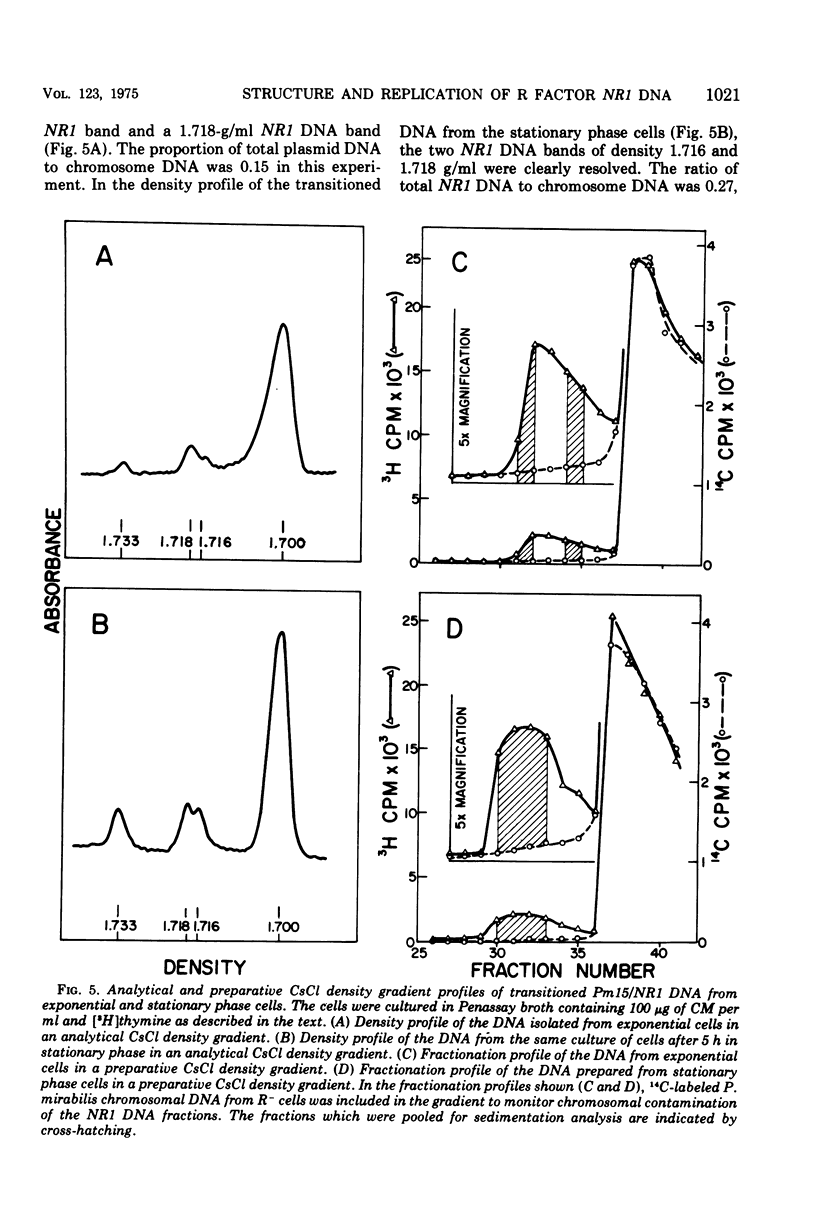
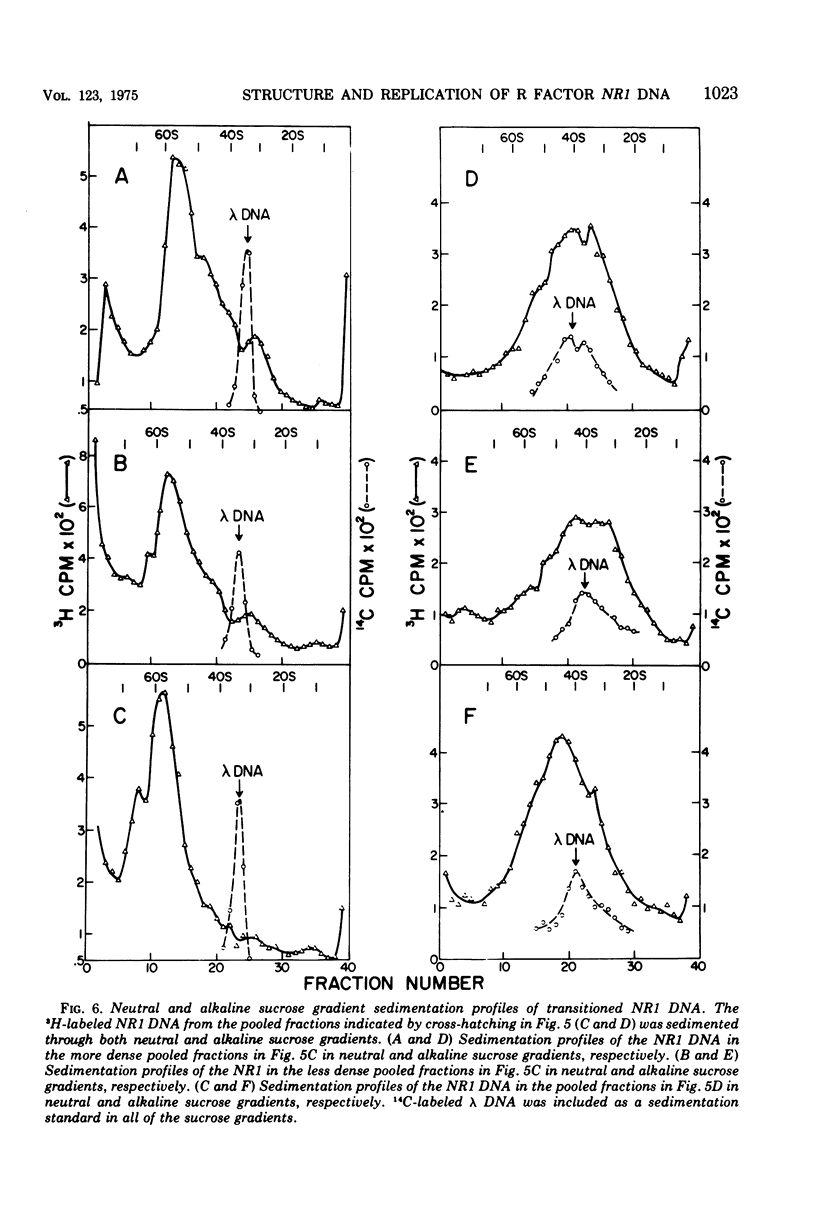
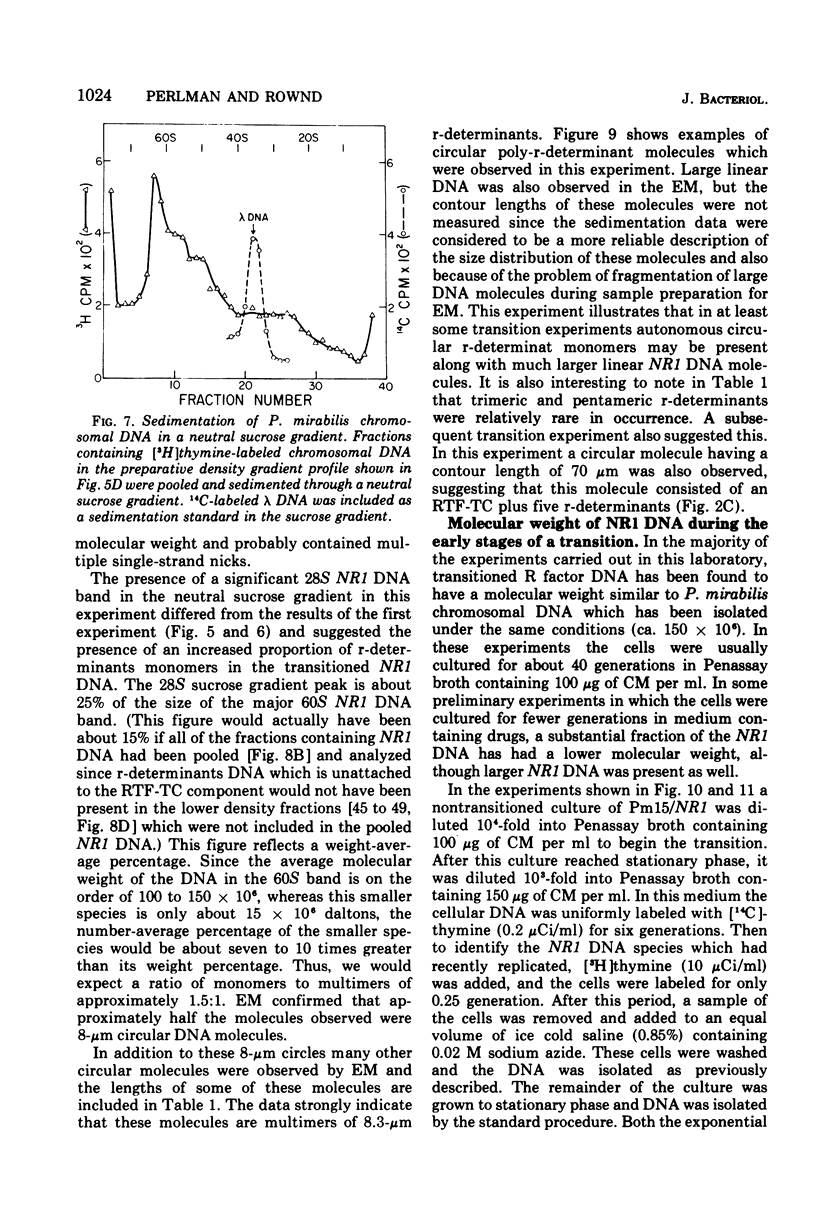
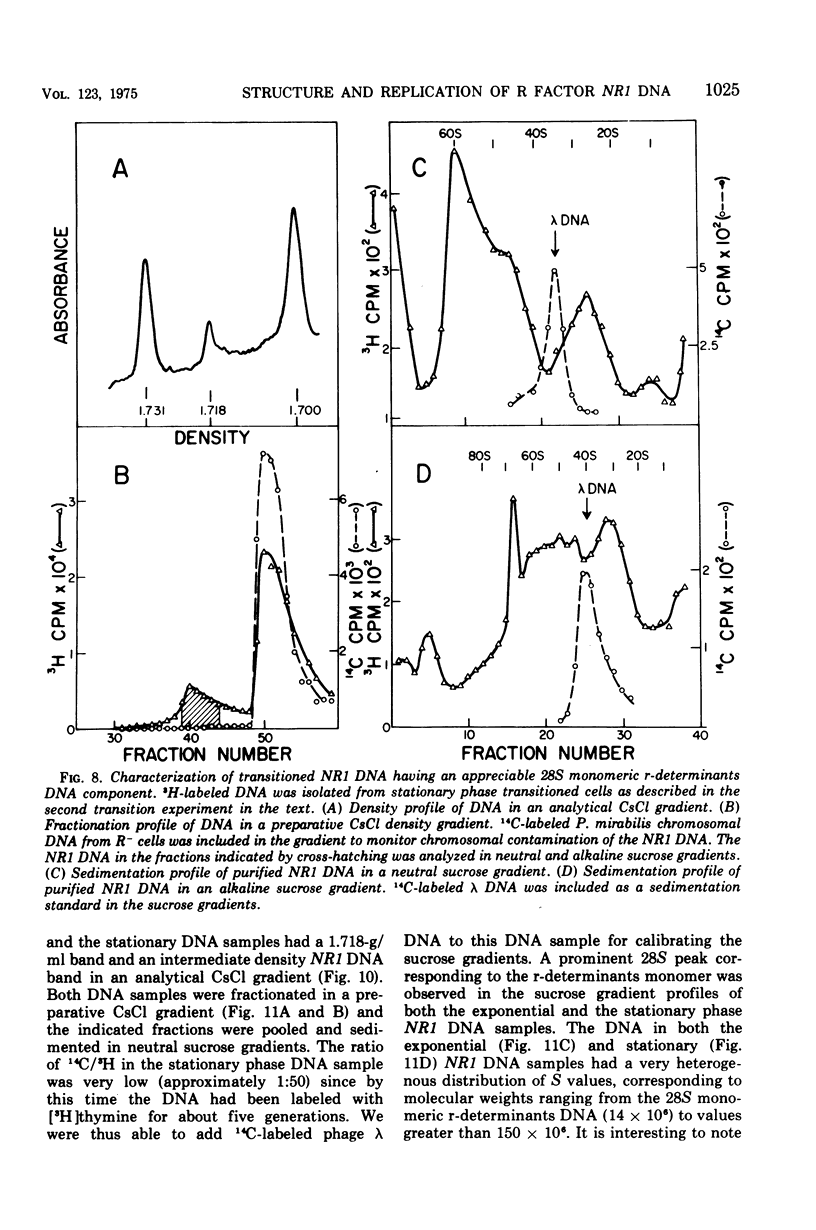
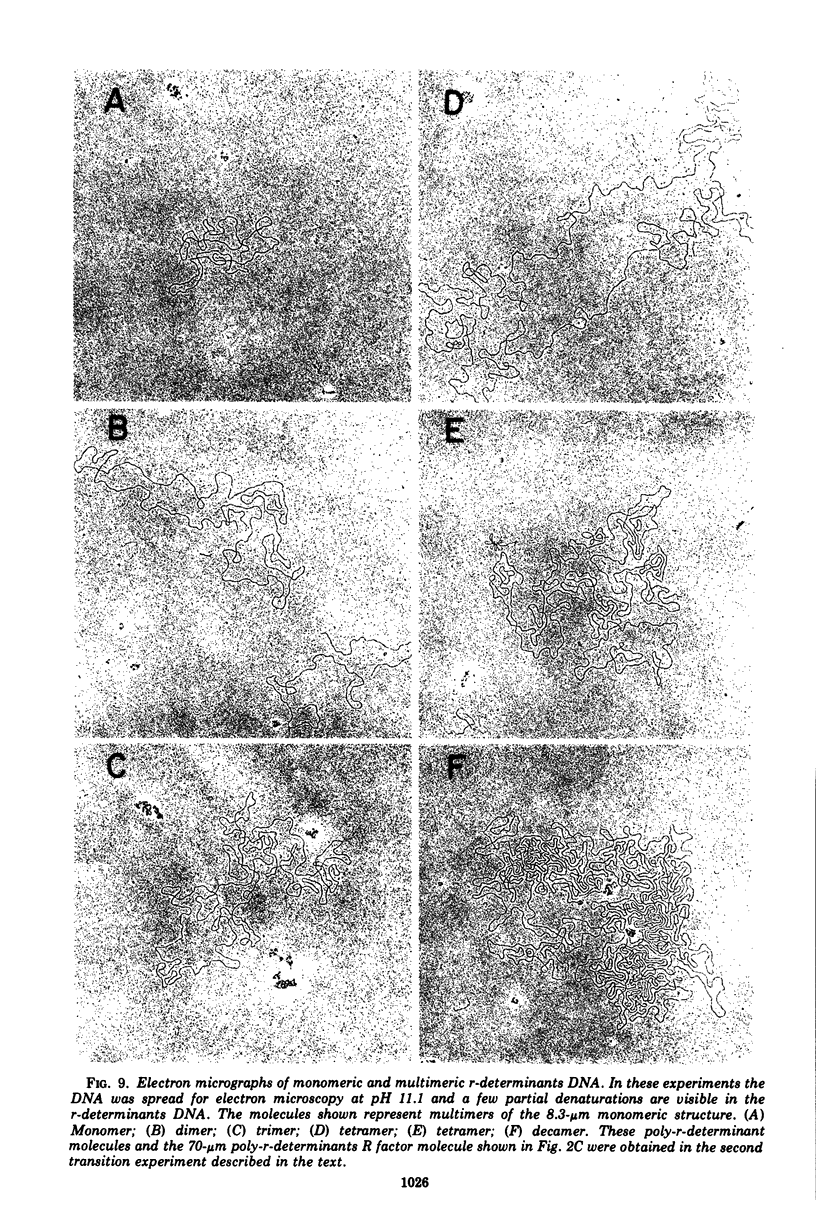
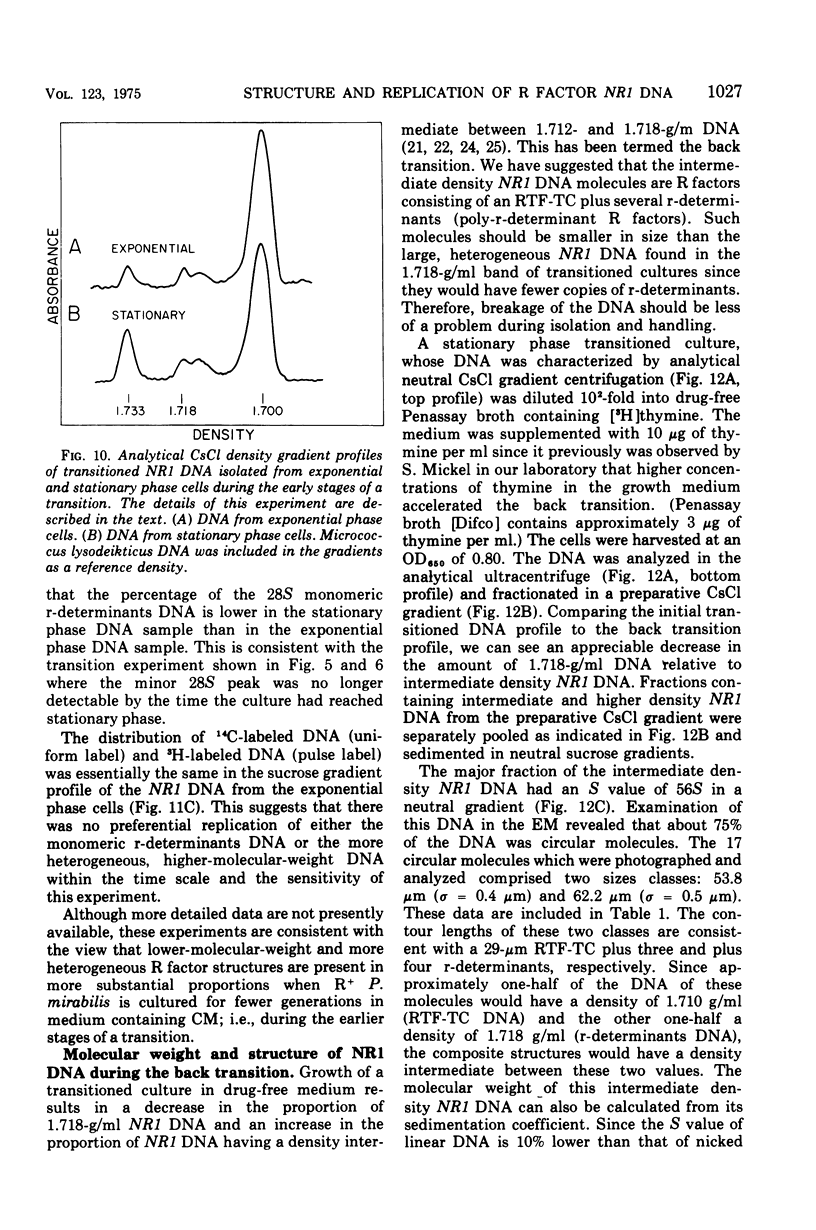
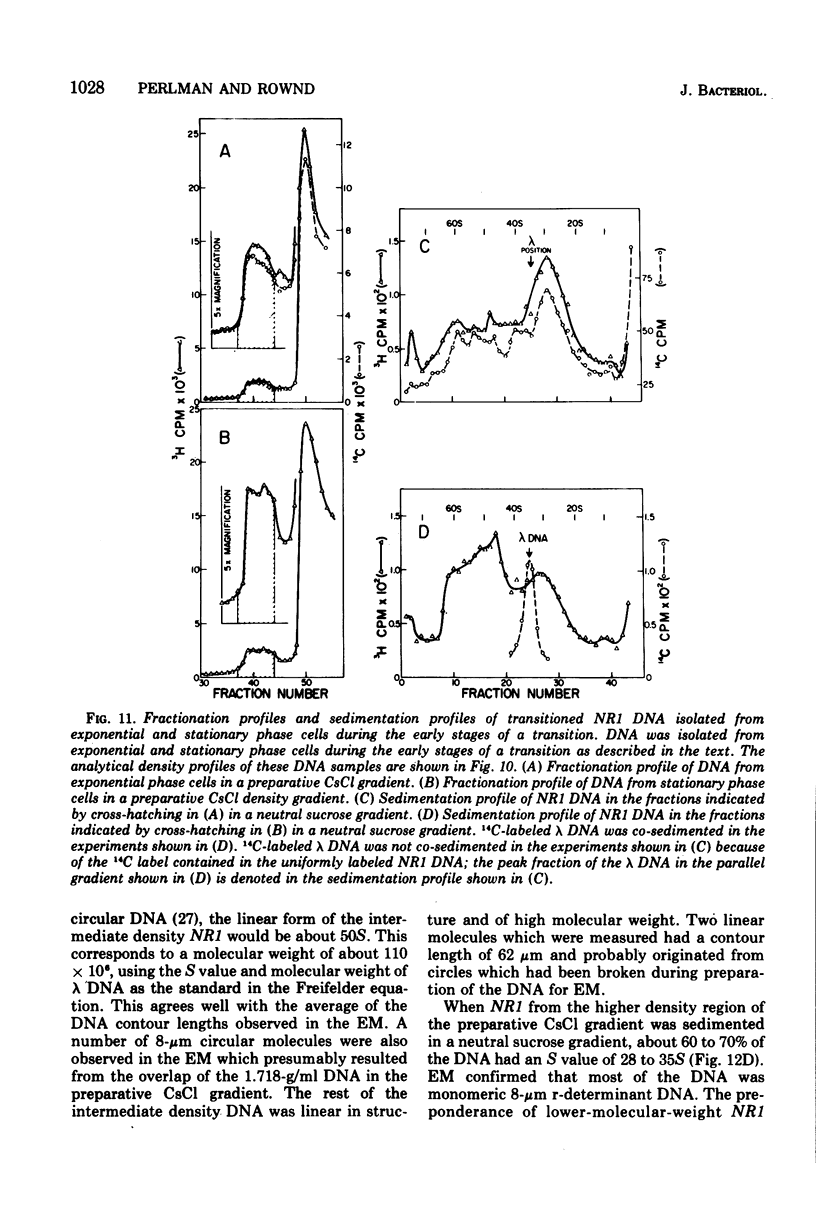
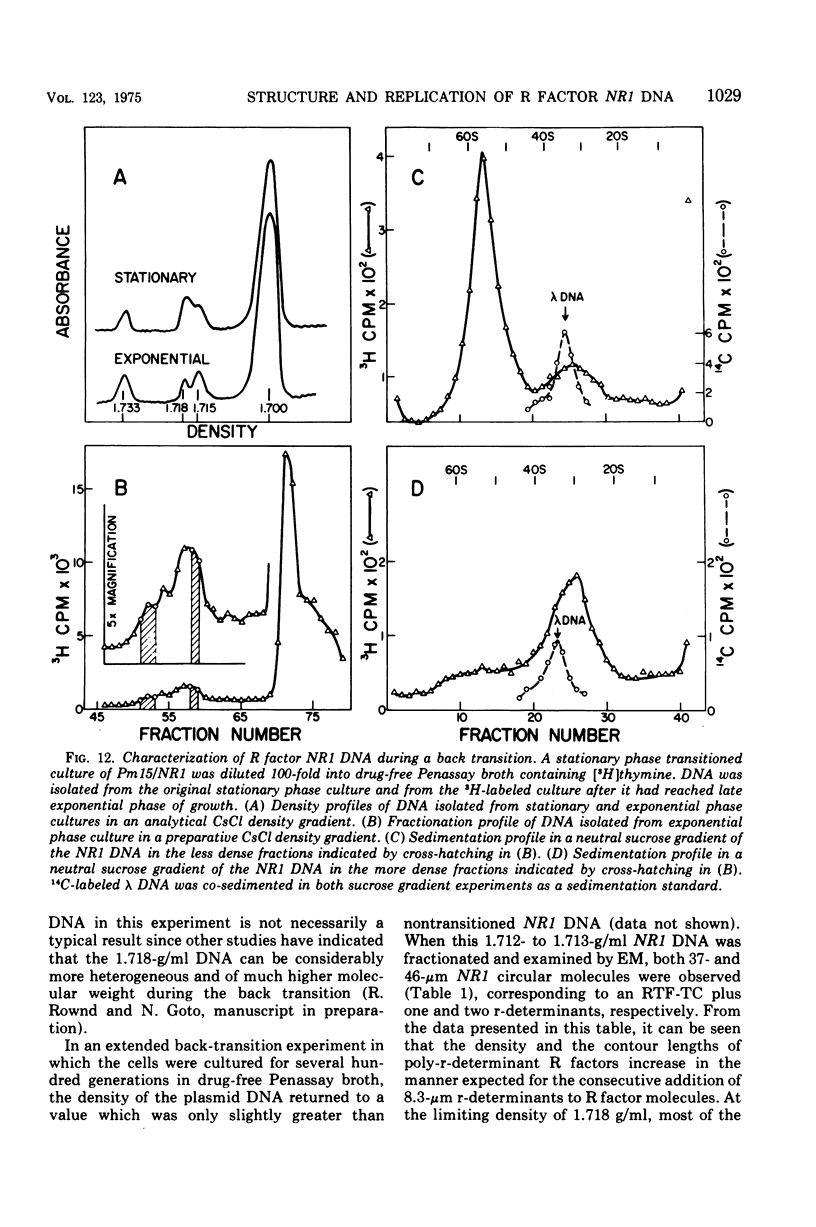
The structure of R factor NR1 DNA in Proteus mirabilis has been studied by using the techniques of CsCl density gradient centrifugation, sedimentation in neutral and alkaline sucrose gradients, and electron microscopy. It has been shown that the nontransitioned form of NR1 DNA isolated from P. mirabilis cultured in drug-free medium is a37-mum circular deoxyribonucleic acid (DNA) with a density of 1.712 g/ml in a neutral CsCl gradient. This circular molecule is a composite structure consisting of a 29-mum resistance transfer factor containing the tetracycline-resistance genes (RTF-TC) and an 8-mum r-determinants component conferring resistance to chloramphenicol (CM), streptomycin/spectinomycin, and the sulfonamides. There are one to two copies of NR1 per chromosome equivalent of DNA in exponential-phase cells cultured in Penassay broth. After growth of PM15/NR1 in medium containing 100 mug of CM per ml, the density of the NR1 DNA increased from 1.712 g/ml to approximately 1.718 g/ml and the proportion of NR1 DNA relative to the chromosome is amplified about 10-fold. The changes in R factor DNA structure which accompany this phenomenon (termed the transition) have been studied. DNA density profiles of the transitioned NR1 DNA consist of a 1.718 g/ml band which is skewed toward the less dense side. The transitioned NR1 DNA consists of molecules containing the RTF-TC element attached to multiple copies of r-determinants DNA (poly-r-determinant R factors) and multimeric and monomeric autonomous r-determinants structures. Poly-r-determinant R factors have a density intermediate between the basic composite structure (1.712 g/ml) and r-determinants DNA (1.718 g/ml). These species presumably account for the skewing of the 1.718-g/ml DNA band toward the less dense side. When transitioned cells are subsequently cultured in drug-free medium, poly-r-determinant R factors and autonomous poly-r-determinants undergo dissociation to form smaller structures containing fewer copies of r-determinants. This process continues until, after prolonged growth in drug-free medium the NR1 DNA returns to the nontransitioned state which consists of an RTF-TC and a single copy of r-determinants.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Miller C. A. Non-chromosomal antibiotic resistance in bacteria. II. Molecular nature of R-factors isolated from Proteus mirabilis and Escherichia coli. J Mol Biol. 1970 Jun 28;50(3):671–687. doi: 10.1016/0022-2836(70)90092-6. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Silver R. P., Sharp P. A., McCoubrey A. E. The problems of drug-resistant pathogenic bacteria. Studies on the molecular nature of R factors. Ann N Y Acad Sci. 1971 Jun 11;182:172–187. doi: 10.1111/j.1749-6632.1971.tb30655.x. [DOI] [PubMed] [Google Scholar]
- Davies J. E., Rownd R. Transmissible multiple drug resistance in Enterobacteriaceae. Science. 1972 May 19;176(4036):758–768. doi: 10.1126/science.176.4036.758. [DOI] [PubMed] [Google Scholar]
- Falkow S., Citarella R. V., Wohlhieter J. A. The molecular nature of R-factors. J Mol Biol. 1966 May;17(1):102–116. doi: 10.1016/s0022-2836(66)80097-9. [DOI] [PubMed] [Google Scholar]
- Freifelder D. Molecular weights of coliphages and coliphage DNA. IV. Molecular weights of DNA from bacteriophages T4, T5 and T7 and the general problem of determination of M. J Mol Biol. 1970 Dec 28;54(3):567–577. doi: 10.1016/0022-2836(70)90127-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helinski D. R. Plasmid determined resistance to antibiotics: molecular properties of R factors. Annu Rev Microbiol. 1973;27:437–470. doi: 10.1146/annurev.mi.27.100173.002253. [DOI] [PubMed] [Google Scholar]
- Hershberger C., Mickel S., Rownd R. Asymmetric distribution of guanine plus thymine between complementary strands of deoxyribonucleic acid of members of the Enterobacteriaceae. J Bacteriol. 1971 Apr;106(1):238–242. doi: 10.1128/jb.106.1.238-242.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Rownd R. Replication of R-factors in Proteus mirabilis: replication under relaxed control. J Mol Biol. 1970 Aug;51(3):473–489. doi: 10.1016/0022-2836(70)90002-1. [DOI] [PubMed] [Google Scholar]
- Morris C. F., Hashimoto H., Mickel S., Rownd R. Round of replication mutant of a drug resistance factor. J Bacteriol. 1974 Jun;118(3):855–866. doi: 10.1128/jb.118.3.855-866.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya R., Rownd R. Transduction of R factors by a Proteus mirabilis bacteriophage. J Bacteriol. 1971 Jun;106(3):773–783. doi: 10.1128/jb.106.3.773-783.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisioka T., Mitani M., Clowes R. C. Molecular recombination between R-factor deoxyribonucleic acid molecules in Escherichia coli host cells. J Bacteriol. 1970 Jul;103(1):166–177. doi: 10.1128/jb.103.1.166-177.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisioka T., Mitani M., Clowes R. Composite circular forms of R factor deoxyribonucleic acid molecules. J Bacteriol. 1969 Jan;97(1):376–385. doi: 10.1128/jb.97.1.376-385.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Twose T. M., Holland M. J., Rownd R. H. Denaturation mapping of R factor deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1035–1042. doi: 10.1128/jb.123.3.1035-1042.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punch J. D., Kopecko D. J. Positive and negative control of R-factor replication in Proteus mirabilis. J Bacteriol. 1972 Jan;109(1):336–349. doi: 10.1128/jb.109.1.336-349.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Kasamatsu H., Mickel S. The molecular nature and replication of drug resistance factors of the Enterobacteriaceae. Ann N Y Acad Sci. 1971 Jun 11;182:188–206. doi: 10.1111/j.1749-6632.1971.tb30656.x. [DOI] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Rownd R., Nakaya R., Nakamura A. Molecular nature of the drug-resistance factors of the Enterobacteriaceae. J Mol Biol. 1966 Jun;17(2):376–393. doi: 10.1016/s0022-2836(66)80149-3. [DOI] [PubMed] [Google Scholar]
- Rownd R., Perlman D., Hashimoto H., Mickel S., Applebaum E., Taylor D. Dissociation and reassociation of the transfer factor and resistance determinants of R factors as a mechanism of gene amplification in bacteria. Johns Hopkins Med J Suppl. 1973;2:115–128. [PubMed] [Google Scholar]
- Rownd R. Replication of a bacterial episome under relaxed control. J Mol Biol. 1969 Sep 28;44(3):387–402. doi: 10.1016/0022-2836(69)90368-4. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J. Physical and topological properties of circular DNA. J Gen Physiol. 1966 Jul;49(6):103–125. doi: 10.1085/jgp.49.6.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE T. Infective heredity of multiple drug resistance in bacteria. Bacteriol Rev. 1963 Mar;27:87–115. doi: 10.1128/br.27.1.87-115.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Ogata Y. Genetic stability of various resistance factors in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1970 May;102(2):363–368. doi: 10.1128/jb.102.2.363-368.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]