Abstract
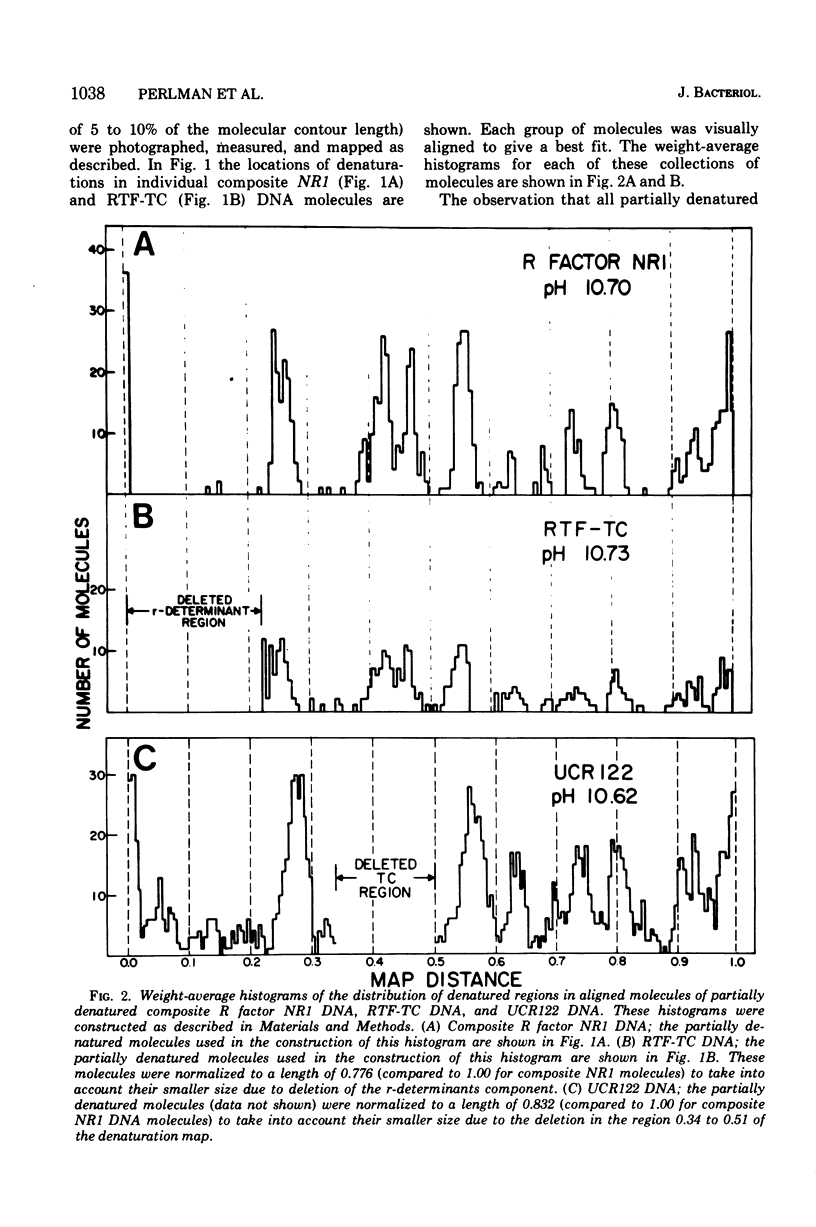
The R factor NR1 consists of two components: a resistance transfer factor which harbors the tetracycline resistance genes (RTF-TC) and the r-determinants component which harbors the other drug resistance genes. Using partial denaturation mapping it is possible to distinguish the RTF-TC region from the r-determinants region of the composite R factor NR1 DNA which has a contour length of 37 mum and a density of 1.712 g/ml. The r-determinants region was a relatively undenatured 8.5-mum segment of the molecule when the deoxyribonucleic acid was partially denatured at pH 10.7. An RTF-TC genetic segregant of NR1 which had lost the r-determinants component had a contour length of 28.7 mum and a density of 1.710 g/ml. Characterization of an RTF-TC using partial denaturation mapping at pH 10.7 confirmed that the relatively undenatured 8.5-mum r-determinants segment of the composite R factor had been deleted. Circular, transitioned NR1 DNA molecules (1.716 to 1.718 g/ml), whose contour lengths were consistent with an RTF-TC plus an integral number of tandem copies of r-determinants, were also characterized by denaturation mapping. The relatively undenatured region in these molecules had a length equal to an integral number of copies of r-determinants and was located at the same site in the partially denatured RTF-TC as the single copy of r-determinants in the 37-mum composite NR1. This indicates that there is a unique integration site for r-determinants in the RTF-TC component. The R factor UCR122, a TC deletion mutant of NR1, was also characterized by denaturation mapping. The translocation of the TC resistance gene(s) on the denaturation map permitted the alignment of the denaturation map with the heteroduplex map of Sharp et al. (u073). Linear and circular monomeric and presumed multimeric r-determinants DNA molecules (p = 1.718 g/ml) were partially denatured at a higher pH (11.10). The r-determinants multimers showed a repeating 8.3-mum (monomeric) partial denaturation pattern indicating a head-to-tail arrangement of monomers in these poly-r-determinant molecules.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Franklin T. J., Rownd R. R-factor-mediated resistance to tetracycline in Proteus mirabilis. J Bacteriol. 1973 Jul;115(1):235–242. doi: 10.1128/jb.115.1.235-242.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Rownd R. H. Transition of the R factor NR1 and Proteus mirabilis: level of drug resistance of nontransitioned and transitioned cells. J Bacteriol. 1975 Jul;123(1):56–68. doi: 10.1128/jb.123.1.56-68.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoar D. I. Fertility regulation in F-like resistance transfer factors. J Bacteriol. 1970 Mar;101(3):916–920. doi: 10.1128/jb.101.3.916-920.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman R. B., Schnös M. Partial denaturation of thymine- and 5-bromouracil-containing lambda DNA in alkali. J Mol Biol. 1970 Apr 14;49(1):93–98. doi: 10.1016/0022-2836(70)90378-5. [DOI] [PubMed] [Google Scholar]
- Perlman D., Rownd R. H. Transition of R factor NR1 in Proteus mirabilis: molecular structure and replication of NR1 deoxyribonucleic acid. J Bacteriol. 1975 Sep;123(3):1013–1034. doi: 10.1128/jb.123.3.1013-1034.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff R., Bauer W., Vinograd J. A dye-buoyant-density method for the detection and isolation of closed circular duplex DNA: the closed circular DNA in HeLa cells. Proc Natl Acad Sci U S A. 1967 May;57(5):1514–1521. doi: 10.1073/pnas.57.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rownd R., Mickel S. Dissociation and reassociation of RTF and r-determinants of the R-factor NR1 in Proteus mirabilis. Nat New Biol. 1971 Nov 10;234(45):40–43. doi: 10.1038/newbio234040a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]


