Abstract
The complete sequence of the Synechocystis chromosome has revealed a phytochrome-like sequence that yielded an authentic phytochrome when overexpressed in Escherichia coli. In this paper we describe this recombinant Synechocystis phytochrome in more detail. Islands of strong similarity to plant phytochromes were found throughout the cyanobacterial sequence whereas C-terminal homologies identify it as a likely sensory histidine kinase, a family to which plant phytochromes are related. An ≈300 residue portion that is important for plant phytochrome function is missing from the Synechocystis sequence, immediately in front of the putative kinase region. The recombinant apoprotein is soluble and can easily be purified to homogeneity by affinity chromatography. Phycocyanobilin and similar tetrapyrroles are covalently attached within seconds, an autocatalytic process followed by slow conformational changes culminating in red-absorbing phytochrome formation. Spectral absorbance characteristics are remarkably similar to those of plant phytochromes, although the conformation of the chromophore is likely to be more helical in the Synechocystis phytochrome. According to size-exclusion chromatography the native recombinant apoproteins and holoproteins elute predominantly as 115- and 170-kDa species, respectively. Both tend to form dimers in vitro and aggregate under low salt conditions. Nevertheless, the purity and solubility of the recombinant gene product make it a most attractive model for molecular studies of phytochrome, including x-ray crystallography.
The Synechocystis chromosome sequence includes an ORF (slr0473) that appears to encode a protein with similarities to phytochrome (1). In earlier reports (2, 3) we noted various features of the sequence showing homology to phytochrome from lower plants and demonstrated by overexpression in Escherichia coli that the ORF indeed codes for a spectrally functional phytochrome.
The discovery of a cyanobacterial phytochrome is significant in various respects. The evolution of a red/far-red photochromic detector has, until now, been considered exclusively within the context of the plant kingdom, whereas if Synechocystis phytochrome represents an ancestral form, the question of its original and subsequent roles in environmental perception must be addressed from a much earlier point in time. The same applies to its biochemical evolution, both in relation to light absorption and signal transduction: the mechanism of plant phytochrome action and its primary reaction partner “X” are unknown. Clues provided by primitive phytochrome systems might be useful here, especially as similarities between plant phytochromes and bacterial sensory kinases have been recognized (4, 5). Since its genome has been sequenced in its entirety, Synechocystis is the first organism in which all the genes involved in a phytochrome signal transduction pathway are known—but have yet to be identified. Conversely, spectrally and biochemically active Synechocystis phytochrome expressed in E. coli might allow potential components of the transduction pathway to be investigated by coexpression in the heterologous host. Finally, the recombinant molecule overexpressed in E. coli offers greatly improved possibilities for studying phytochrome molecular structure. Not only is the preparation of large amounts of pure apoprotein facile, the product is highly soluble and readily attaches its chromophore to form spectrally active holoprotein (3).
In this paper, we describe Synechocystis phytochrome in more detail, in particular its sequence homologies, and the spectral and biochemical characteristics of the recombinant protein from E. coli.
MATERIALS AND METHODS
Cloning Procedures.
The Synechocystis sp. PCC 6803 ORF slr0473 (phy) was amplified from genomic DNA by PCR using error-checking polymerase (Vent; Biolabs, Northbrook, IL). Primers used were designed for efficient ligation and expression with the vector pQE12 (Qiagen, Chatsworth, CA). The N-terminal primer was 5′-GGAATTCATTAAAGAGGAGAAATTAACTATGGCCACCACCGTACAACTCAG, designed to incorporate an EcoRI site and a Shine–Dalgarno sequence upstream of the template ORF (bold). For the pF10.His clone the C-terminal primer was 5′-GGGAGATCTGTTGCCAATGGGGATGGAGAAG; with this, a BglII site was inserted downstream of the template ORF (bold), omitting the stop codon. This provided a C-terminal translational fusion to six histidine codons from the vector, and hence a convenient affinity tag. A second clone, pF10.Stop, was generated as above but with the C-terminal primer 5′-GGGGATCCTTAGTTGCCAATGGGGATGGAG; here the native stop codon of the ORF template (bold) was retained and a BamHI site attached downstream. The PCR profile was as follows: 3 times each at 94°C for 1 min, 62°C for 1 min, and 68°C for 2 min; 30 times each at 94°C for 1 min and 68°C for 3 min; 1 time at 68°C for 10 min, all with a 100 pg genomic DNA template. The PCR products were ligated into the vector and cloned in XL1-Blue (Stratagene), selecting with 100 μg/ml ampicillin and 30 μg/ml tetracycline (Biomol, Hamburg, Germany) according to standard procedures (7) (all enzymes from New England Biolabs). Coexpression with the E. coli chaperonins GroES and GroEL was achieved using the compatible plasmid pGroESL (a generous gift of G. Lorimer, Du Pont) and additionally selecting with 70 μg/ml chloramphenicol (8).
Expression and Purification of Apoprotein.
Liquid RB (1% tryptone/0.5% yeast extract/0.5% NaCl/0.2% glucose, pH 7.6/NaOH, plus antibiotics) cultures up to a 2 liter volume were grown with vigorous shaking at 30°C to OD550 nm 0.5, then transferred to 20°C for 1 h before induction with 1 mM isopropyl β-d-thiogalactoside (Biomol) for 10–14 h. After that time, the phytochrome apoprotein had accumulated to ≈50% of the total protein. The cells were then pelleted at 5,000 × g for 5 min at 4°C, washed in 50 mM Tris⋅Cl (pH 7.8), 5 mM EDTA, 300 mM NaCl, pelleted again, and resuspended in 1/10th the original culture volume of 50 mM Tris⋅Cl pH 7.8, 300 mM NaCl, 5 mM EDTA, and 1 μg/ml lysozyme (Biomol) before freezing at −80°C. The slurry was thawed slowly then sonicated (Branson B15) at 4–15°C until no further protein was released. Debris was pelleted and ammonium sulfate (AmS) added to the supernatant from a buffered 3.3 M stock to give a final concentration of 1.3 M. The protein was precipitated at 4°C and pelleted at 20,000 × g.
Pellets from the histidine-tagged clones were suspended in WB [50 mM Tris⋅Cl, pH 7.8/300 mM NaCl/5 mM imidazole (Sigma)], recentrifuged, and the supernatant (protein concentration 1–5 mg/ml) applied to a WB-equilibrated nickel affinity column (Bio-Rad Econosystem with Qiagen Ni2+-NTA agarose matrix, diameter 1.4 cm, height 12 cm, flow rate 4 ml/min). The apoprotein bound quantitatively to the matrix. After washes with 200 ml WB and 100 ml salt-free buffer, apoprotein was eluted in essentially pure form with 250 mM imidazole in 50 mM Tris⋅Cl (pH 7.8), as shown (3). The eluate was concentrated via AmS precipitation and taken up in buffer.
Protein concentrations were derived from Bradford assays calibrated against BSA standards and from carefully weighed milligram amounts of pure recombinant apoprotein, lyophilized after extensive dialysis against water. Extinction coefficients were calculated from spectral absorbance data based on such preparations.
Preparation of Holoprotein.
Manipulations were carried out under a green safelight. Phycocyanobilin (PCB) was prepared by methanolysis of Spirulina geitlerie followed by HPLC (9) and crystallization (see ref. 6). Concentrated solutions in dimethyl sulfoxide were stored at −20°C in darkness. PCB was quantified spectrophotometrically in methanol-HCl (19/1) from ɛ680 = 3.7 × 104 M−1⋅cm−1 (10). Phytochromoblin (PΦB) was prepared according to Cornejo et al. (11) and stored in dimethyl sulfoxide in darkness.
Autoassembly with PCB was investigated with a 400 μl 16 μM PCB aliquot in 50 mM Tris⋅Cl (pH 7.8), 5 mM EDTA buffer in a quartz cuvette, allowing A250–800 spectra to be measured (Uvikon 941, Milan) during the reaction. Apoprotein prepared as above was added as a 100 μl, 5 mg/ml aliquot and mixed rapidly in situ. Spectra were recorded immediately and after assembly was complete. Alternatively, absorbance was monitored continuously at 660 or 700 nm and covalent attachment monitored as described below. The mixing procedure gave a small excess of chromophore substrate relative to the phytochrome apoprotein. In the case of the pF10.Stop clone, the crude AmS pellet was resuspended in buffer at 5 mg/ml. As PΦB was available in limited amounts, a small excess of apoprotein was used. Excess PCB was removed from holoprotein by AmS precipitation or G25 Sephadex gel filtration (Nap10, Pharmacia).
Open-chain tetrapyrroles bind Zn2+, forming a helix wrapped tightly around the hydrated ion (12). This was exploited in two ways. First, Zn2+-complexed linear tetrapyrroles and their adducts fluoresce orange in UV/B (13). Thus, covalent attachment of PCB to the apoprotein was detected via SDS/PAGE (14) followed by incubation of the gel in 1 mM zinc acetate for 15 min and UV/B fluorography (TM20 transilluminator and GDS5000 with Enhanced Analysis SYstem; Ultraviolet Products, San Gabriel, CA). Attachment kinetics were measured by rapidly injecting aliquots into boiling SDS-sample buffer at different time points after mixing apoprotein and PCB. Second, the charge carried by the complexed ions retards migration (15), allowing apoprotein and holoproteins to be distinguished following conventional Coomassie staining. Thus, in zinc–SDS/PAGE, zinc acetate at 1 mM was included in all components of the Laemmli gel system during electrophoresis.
Spectral Analysis.
Holoprotein preparations were placed in a quartz cuvette with polished side walls in a Uvikon 941 spectrophotometer modified to allow absorbance measurements during photoconversion in situ. Red (R, 658 ± 10 nm) and far-red (FR, 730 ± 12 nm; type DAL interference filters, Schott, Mainz, Germany) collimated actinic light was provided unilaterally at 10 μmol m−2⋅s−1 from a projector. Spectra were recorded 250–800 nm after saturating irradiations. Red-absorbing phytochrome/far-red-absorbing phytochrome (Pr ↔ Pfr) photoconversion kinetics were monitored at 658 or 730 nm. Photoconversion rates were essentially first order during the first minute of irradiation and were used to derive [Pfr]max, 658 from Butler’s model (16, 17) allowing the Pfr spectrum to be calculated from the raw absorbance data.
Native Conformation.
The native conformation of apo- and holoproteins was analyzed via a Superose 6 size-exclusion column (Pharmacia) calibrated with six proteins of known molecular size between 12.4 and 443 kDa (Sigma). The column was run at 500 μl/min with 10 mM Tris⋅Cl (pH 7.8), 150 mM NaCl, and 1 mM EDTA. Samples were loaded via a 100 μl loop. A280 was monitored and 500 μl fractions collected. Native PAGE was also used in this context. Gels of 7.5% acrylamide, 0.2% bisacrylamide were cast with 375 mM Tris⋅Cl (pH 8.8). The running buffer was 180 mM glycine, 25 mM Tris-base. Samples were loaded in 50 mM Tris⋅Cl, 5 mM EDTA pH 7.8 with 10% glycerol and run out at 30 V/cm, then stained with Coomassie. The calibration proteins were BSA monomer and dimer (66 and 132 kDa, respectively) and ovalbumin monomer and dimer (45 and 90 kDa, respectively), as recommended by Sigma.
RESULTS
Homology.
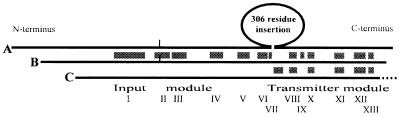
A conservative alignment of Synechocystis phytochrome [CyanoBase slr0473; (1)] with phytochrome from the moss Ceratodon purpureus [Cerpu_PHY_2, GenBank accession no. U56698 (2)] on the one hand and the RcsC osmosensory histidine kinase from E. coli [GenBank accession no. D28242 (18)] on the other was derived using macaw, blastp (National Center for Biotechnology Information, Bethesda, MD) and clustalw (19). Thirteen islands of close similarity between the moss and cyanobacterial sequences were recognized (Fig. 1 and Table 1). Additional, weaker homologies (not shown) are also recognizable in the extreme N terminus. Islands I–VII show clear homologies between the phytochrome N termini. The C-terminal region of Synechocystis phytochrome shows homologies both to the sensory kinase and to the moss phytochrome. On the other hand, the phytochrome alignment requires an ≈300-residue gap in the Synechocystis sequence, immediately preceding the first homologies to sensory kinases.
Figure 1.
Associations of Synechocystis phytochrome to plant phytochromes and bacterial sensor proteins. Lines: A, Lower plant phytochrome (Cerpu_PHY0_2, U56698, from the moss Ceratodon purpureus; 1,121 residues); B, Synechocystis phytochrome (Phy, slr0473; 748 residues); and C, histidine kinase sensor protein (RcsC, M28242, from E. coli; 933 residues). The putative chromophore attachment site is marked (|) in lines A and B. Synechocystis phytochrome shows islands (subdomains I–XIII, shaded; see Table 1) of strong similarity to plant phytochromes at intervals throughout its length, although 306 residues of the plant phytochrome apoprotein (E563..S869 in Cerpu_PHY0_2) are missing from the Synechocystis sequence.
Table 1.
Ceratodon phytochrome and RcsC relative to Synechocystis phytochrome: percent amino acid identities (and similarities, in parentheses) in the homologous islands (subdomains I–XIII) in Fig. 1
| Subdomain | Comment | Residues in Synechocystis phytochrome | Ceratodon phytochrome Cerpu PHY_2, % | RcsC, % |
|---|---|---|---|---|
| I | Phosphotyrosine motif | D158..D226 | 62 (86) | |
| II | Chromophore | V246..D280 | 44 (67) | |
| III | L283..S314 | 56 (75) | ||
| IV | Q366..L395 | 37 (67) | ||
| V | G425..G452 | 43 (61) | ||
| VI | L469..V492 | 50 (71) | ||
| VII | E493..L500 | 50 (88) | ||
| VIII | H | S537..E555 | 35 (71) | 41 (76) |
| IX | L562..D571 | 50 (80) | ||
| X | S578..V593 | 13 (56) | 44 (75) | |
| XI | N | V635..A657 | 39 (74) | 43 (78) |
| XII | G1 + F | F677..T703 | 15 (33) | 33 (56) |
| XIII | G2 | G709..I721 | 38 (77) | 77 (92) |
Subdomain I includes the perfectly conserved motif EPYLGLHY, resembling the mammalian phospholipase C γ binding site. Subdomain II resembles the consensus chromophore binding region of plant phytochromes and includes the invariate CH at the point of chromophore attachment. The equivalent H (histidine autophosphorylation site), N, G1, F and G2 subdomains described by Parkinson (25) are indicated. The alignment on which this analysis is based is available from the authors.
Autoassembly.
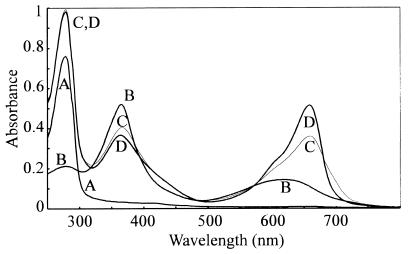
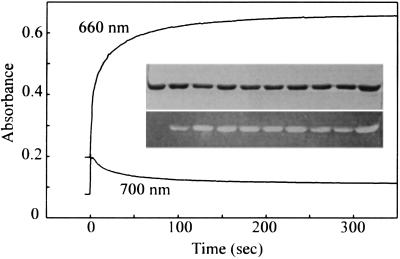
The recombinant Synechocystis PHY apoprotein overexpressed in E. coli could be purified to homogeneity by a single affinity step (3). Spectral absorbance changes accompanied the uptake of PCB by the apoprotein (Fig. 2), allowing self-assembly to be monitored continuously (Fig. 3). Absorbance at 370 nm fell whereas that at 660 and 700 nm increased dramatically within 10 seconds of mixing the apoprotein. Thereafter, whereas A660 continued to increase slowly, reaching a constant value after about 10 min, A700 fell asymptotically over a similar period. Zn2+-induced fluorescence of tetrapyrrole protein adducts in SDS gels indicated that covalent attachment of the chromophore was largely complete within 10 s (Fig. 3 Inset).
Figure 2.
Autoassembly with PCB. Changes in absorption spectra during autoassembly. Curves: A, apoprotein; B, PCB; C, 10 s after mixing; and D, 1,000 s after mixing. Absorbances in curves A and B have been arithmetically corrected for dilution to allow quantitative comparison.
Figure 3.
Assembly kinetics of Synechocystis phytochrome. Spectrophotometric measurements at 660 and 700 nm. Final concentrations of PCB and apoprotein were 16 μM. (Inset) SDS/PAGE of PHY apoprotein (t = 0) and PCB–phytochrome adducts during assembly. (Upper) Coomassie-stained gel. (Lower) Zn2+-induced tetrapyrrole fluorescence in UV/B. Lanes (left to right) show samples taken at t = 0, 5, 10, 20, 40, 60, 90, 120, 240, 480 s after mixing.
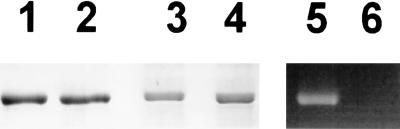
In normal SDS/PAGE, holoproteins and apoproteins migrated identically (Fig. 4, lanes 1 and 2, respectively), whereas in zinc–SDS gels, the holoprotein was retarded slightly with respect to the apoprotein (lanes 3 and 4, respectively), consistent with zinc binding to the chromophore and reducing the net charge of the SDS–holoprotein complex (15).
Figure 4.
Coomassie staining (lanes 1–4) and Zn2+-induced tetrapyrrole fluorescence under UV/B (lanes 5–6). Lanes 1 and 2; SDS/PAGE, lanes 3–6; zinc–SDS/PAGE. Lanes 1, 3, and 5; holoprotein (assembled with PCB); lanes 2, 4, and 6; apoprotein (without PCB).
Spectral Characteristics.
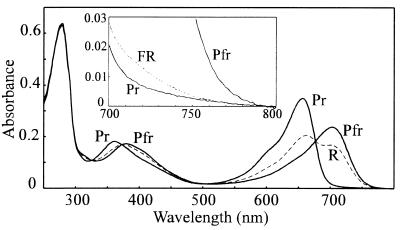
R/FR photochromicity of the PCB holoprotein adduct has been described (3). The true Pfr and Pr absorbance spectra are shown in Fig. 5. [Pfr]658 (the mol fraction of Pfr at photoequilibrium in R) was derived from initial rates of photoconversion to be 0.68 ± 0.02. The Pfr spectrum was calculated from the raw spectra following saturating R and FR via this factor. The far-red peak at 702 nm was clearly resolved, and little trace of a shoulder at 658 nm, the Pr maximum, remained. The Pr spectrum shown is for holoprotein self-assembled in total darkness; the spectrum following R/FR cycling was significantly different. The FR—dark difference spectrum (data not shown) indicated that the effect was due to the significant absorbance of Pr in the far-red region (Fig. 5 Inset) establishing a [Pfr]730 of ≈0.05. Absorption spectra following R remained stable in darkness for several hours (data not shown), indicating that Pfr dark reversion and decay were negligible.
Figure 5.
Spectral absorbance characteristics of PCB adducts in Pr and Pfr forms. The Pr spectrum was measured following autoassembly in total darkness; the Pfr spectrum was derived from raw data for the photoequilibrium mixture in R (dashed line), using [Pfr]max, 657 = 0.68 calculated from initial phototransformation rates. (Inset) Far-red absorbance of unirradiated sample (Pr) and following R then FR irradiation (dashed line). Pfr is included for comparison (10-fold expanded scale).
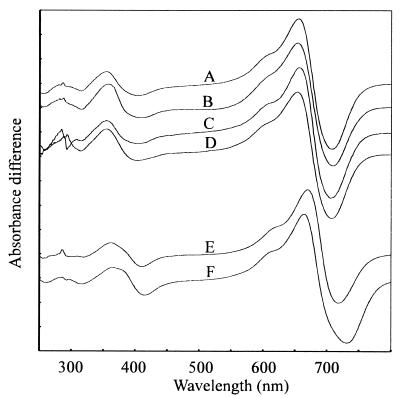
The FR—R difference spectrum for PCB-conjugated pF10.His recombinant phytochrome following affinity purification indicated maxima at 655 and 708 nm and an isosbestic point at 677 nm (3) (Fig. 6, curve A). Difference spectra for various holoprotein preparations were compared with this. Similar data were obtained for putative monomer purified via size-exclusion chromatography, for preparations from apoprotein coexpressed with GroES and GroEL chaperonins, and for crude preparations of wild-type Synechocystis phytochrome lacking the polyhistidine affinity tag (Fig. 6, curves B–D, respectively). However, the PΦB adduct showed a clear red shift with difference peaks at 670 and 719 nm and an isosbestic point at 680 nm (Fig. 6, curve E). The equivalent values for oat phyA prepared according to Grimm and Rüdiger (22) were 664, 732 and 688 nm, respectively (Fig. 6, curve F).
Figure 6.
Synechocystis and oat phytochrome difference spectra. Curves: A, pF10.His-PCB adduct, eluate from affinity column; B, pF10.His-PCB adduct, purified monomer from size-exclusion column; C, pF10.His-PCB adduct, coexpressed with chaperonins; D, pF10.Stop-PCB adduct, AmS-precipitated bacterial lysate; E, pF10.His-PΦB adduct, eluate from affinity column; and F, phytochrome from etiolated oat. The curves have been arithmetically normalized and offset for clarity.
Native Conformation.
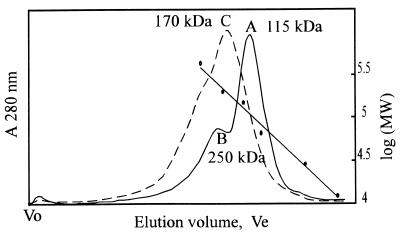
The predominant apoprotein species migrated as a 110- to 115-kDa peak according to size-exclusion chromatography (Fig. 7, curve A). The holoprotein consistently eluted somewhat earlier, corresponding to a 150- to 180-kDa species (Fig. 7 curve C). The apoprotein also showed a pronounced presumptive dimer peak eluting as ≈250 kDa (Fig. 7, curve B). Indeed both apoproteins and holoproteins tended to aggregate, especially at lower pH and salt concentrations (data not shown).
Figure 7.
Size-exclusion chromatography elution profile (Superose 6) of Synechocystis PHY apoprotein (solid curve) and holoprotein (dashed curve) and calibration. Presumptive monomer (curve A) and dimer (curve B) migrated as 115- and 250-kDa peaks, respectively; the holoprotein migrated as 170 kDa (curve C). Calibration points were cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), BSA (66 kDa), alcohol dehydrogenase (150 kDa), β-amylase (200 kDa), and apoferritin (443 kDa). Void volume (Vo) was measured with blue dextran.
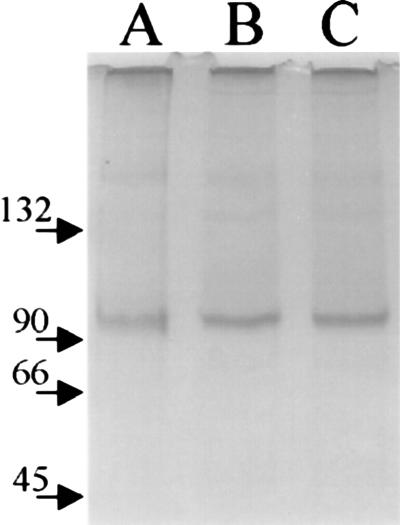
On nondenaturing PAGE gels (Fig. 8) apoproteins and holoproteins showed an effective molecular size corresponding to 95–100 kDa in relation to the marker proteins, the holoprotein migrating only marginally more slowly than the apoprotein. There was little evidence of dimer formation under these conditions, whereas high molecular weight aggregates were apparent. This effect is consistent with size-exclusion profiles run at similarly low salt concentrations.
Figure 8.
Native PAGE of phytochrome apoprotein (lane A) and holoprotein in Pr (lane B) and Pfr (lane C) forms. The position of marker proteins is indicated.
DISCUSSION
The Synechocystis ORF slr0473 shows homologies to phytochrome throughout its length (Fig. 1 and Table 1). In particular, the chromophore-binding region, strongly conserved in all plant phytochromes, can be recognised clearly (subdomain II). This contrasts with the “phytochrome-like” RsaE (23) and PlpA (24) gene products in which such sequences are conspicuously absent (23). Although both are involved in photoperception, there is currently no evidence that either is a photoreceptor.
The C-terminal homologies to RcsC (subdomains VIII and X–XIII, Fig. 1 and Table 1) and numerous other sensory histidine kinases [data not shown; see ref. 25] imply that Synechocystis phytochrome is indeed a member of that family and further reinforces the relationship between phytochromes and sensory histidine kinases first recognised by Schneider-Poetsch (4, 5). Sensory kinases interact with response regulators in “two-component” signaling systems in bacteria and eukaryotes (25–27). Sensory kinases consist of an input module attached to a transmitter module bearing a conserved autophosphorylated histidine residue. Response regulators consist of a receiver module bearing a conserved aspartate residue targeted by the kinase, and an output module. As they commonly act as environmental probes, many input modules span the plasmamembrane; others are cytoplasmic. The input module reacts to changes in specific conditions, regulating kinase activity. This in turn regulates the phosphorylation state and hence output activity of the response regulator. The presumptive phosphohistidine involved in Synechocystis phytochrome is H538 in subdomain VIII. Interestingly, immediately downstream of the phy gene in an operon-like configuration lies a second ORF (slr0474) showing homologies to response regulators. By analogy to CheY (25), the likely aspartate phosphorylation target D68 forms an acid pocket with E15 and D16. It is tempting to speculate that this protein might represent the response regulator with which the phytochrome interacts in Synechocystis.
However, it is difficult to argue that plant phytochromes and sensory kinases function analogously. For example, although the autophosphorylation subdomain is clearly represented in most plant phytochrome sequences (Fig. 1 and Table 1, subdomain VIII), the crucial autophosphorylated H residue is often replaced by R or Q (4). Furthermore, although an appropriate H residue is conserved in some A-type plant phytochromes, physiological activity is retained in H → L mutants (28). The same authors showed that other residues analogous to those considered important in sensory kinases can be mutated without losing physiological function. None of the 17 functionally essential residues in PHYA and PHYB are in sensory kinase-homologous subdomains (29). It would thus seem unlikely that the bacterial sensory kinases provide a simple model for plant phytochrome signal transduction.
The portion of plant phytochrome “missing” from the Synechocystis phytochrome (Fig. 1) is of particular interest. An ≈1-kb insertion would seem to have occurred during the evolution of plant phytochromes from a prokaryotic ancestor. Intron no.3 in Cerpu_PHY_2 is positioned exactly at the 3′ end of the putative insertion. Functionally, the additional sequence lies exactly between the input (phytochrome specific) and transmitter (kinase-like) modules, spanning the so-called “hinge” direct repeat in plant phytochromes (30), including the almost invariant phytochrome LIPPIF motif and the epitopes of both Z-3B1 and P-25 “universal” anti-phytochrome mAbs. Further, 12 of the 17 residues known to be functionally essential in plant phytochromes are located in this part of the molecule (29).
The missing region is also of interest regarding phytochrome quarternary structure. Higher plant phytochrome is an ellipsoidal dimer in its native state: the subdomain responsible for cohesion (33) is in the region missing from Synechocystis phytochrome. Indeed, size exclusion chromatography data (Fig. 7) imply that the native apoprotein exists predominantly as an ellipsoidal monomer, migrating as ≈115 kDa, whereas the dominant holoprotein species behaves as an ≈170-kDa species. Putative dimers do tend to form under the salt conditions normally used with Superose (here 150 mM NaCl), whereas in buffers of low ionic strength, aggregation is seen. This is unavoidable in native PAGE (Fig. 8) where NaCl must be omitted: here the molecular size of the putative monomers was 95–100 kDa relative to calibration standards. In accordance with their role in environmental perception, most sensor histidine kinases are membrane proteins and thus require detergent treatment for solubilization in aqueous buffers, a procedure which abolishes most quarternary structure. Soluble sensor histidine kinases include CheA (in which the sensor module itself is a separate, membrane-bound protein) and NtrB. According to partner-swapping studies using differently marked subunits and size-exclusion chromatography, the dissociation constant for CheA dimers is about 0.3 μM (31) and much lower for NtrB (32). It seems that all sensor histidine kinases require dimerization for activity, at least partly because the histidine “autophosphorylation” occurs in trans between subunit partners (25). We are studying the molecular interactions of Synechocystis phytochrome in this context.
We studied the autoassembly and spectral characteristics of Synechocystis phytochrome in some detail. That these are strikingly similar to those of plant phytochromes, in harmony with homologies in the N-terminal region (subdomains I–VII, Fig. 1 and Table 1), contrasts with the apparent C-terminal differences discussed above.
The capacity of the Synechocystis PHY apoprotein to autoassemble with linear tetrapyrroles to form photochromic holoprotein adducts is typical of phytochrome (34). We monitored this process via Zn2+-induced fluorescence in SDS/PAGE and simply by recording absorbance changes (Figs. 2 and 3). Our data are consistent with that for plant phytochromes expressed in yeast (35, 36) but additionally shows that autoassembly of Synechocystis phytochrome occurs in at least two kinetically distinct phases. Covalent attachment of the chromophore is exceedingly rapid (<10 s) and was associated with a dramatic increase in both A660 and A700. This is followed by a slower phase during which A660 increases further, whereas A700 falls, presumably resulting from conformation changes in the nascent holoprotein. Holoprotein formation is associated with an increase in effective hydrodynamic radius (Fig. 7). The data calls into question the assumption that phytochrome apoprotein is equivalent to Pr. Similar measurements with plant phytochrome apoprotein and holoprotein are currently being carried out (A. Remberg and W.G., unpublished data).
The spectral characteristics of Synechocystis phytochrome are remarkably similar to those of plant phytochromes (Figs. 5 and 6); the same is true for Raman spectroscopic data (see ref. 6). The absorbance maxima and isosbestic point (658, 702, and 677 nm, respectively) of the PCB adduct (Fig. 6, curves A–D) are all blue-shifted with respect to phytochrome purified from higher plants, in which the chomophore is PΦB. Such a blue shift is also observed when higher-plant phytochrome autoassembles with PCB. Reported difference maxima for such adducts are at 650–658 and 709–720 nm (11, 34–41); the equivalent values for our preparations are 655 and 708 nm. Conversely, the difference spectrum of our PΦB adduct is similar to that of oat PHYA (Fig. 6, curves E and F). Although PCB is abundant in the cyanobacterial cytoplasm, we have no evidence that it is the native chromophore: we intend to purify phytochrome from Synechocystis itself to clarify this. Kidd and Lagarias (42) suggested that phytochrome from the alga Mesotaenium might be a PCB rather than a PΦB adduct.
Using Butler′s model (16) [Pfr]658, max was calculated to be 0.68, a low value in harmony with the prominent R shoulder and poorly resolved Pfr maximum in the absorbance spectrum at photoequilibrium in R (3). This mol fraction allowed the Pfr spectrum to be derived (Fig. 5). The significant absorbance of Pr in FR leads to a [Pfr]730 mol fraction of about 0.05, accounting for the difference between the absorbance spectrum of the holoprotein autoassembled in total darkness (pure Pr) and that after FR irradiation (Fig. 5 Inset).
We were able to determine molar extinction coefficients for Synechocystis phytochrome on the basis of absorbance measurements and direct weighings of the pure, salt-free apoprotein. The ɛmax, 280 of 1.0 × 105 M−1⋅cm−1 is similar to that determined for plant phytochromes (43). This is to be expected, because, although the latter have a significantly higher molecular mass (≈124 kDa vs. 88 kDa), the number of aromatic amino acids per molecule is similar. The Butler model allows the overall quantum efficiencies, φ, for Pr → Pfr and Pfr → Pr to be calculated from extinction coefficients of Pr and Pfr and initial photoconversion rates in each direction. We compute similar φ values of ≈0.16 for both: this is similar to estimates for plant phytochromes (43).
The ratio of the Pr absorbance maximum to the 280 nm peak (SAR) has often been used as a parameter of phytochrome purity, homogeneous oat PHYA having an SAR close to unity. The equivalent 658/280 nm SAR for our preparations is about 0.55. This low value is likely to be characteristic of the holoprotein, at least for that expressed in E. coli. Holoprotein preparations purified by size-exclusion chromatography to exclude possible contaminants invisible in SDS/PAGE showed a similar SAR of 0.6. The C-terminal histidine tag had no effect on the spectral properties of the holoprotein (Fig. 6, curves A and D). Zinc–SDS/PAGE (Fig. 4) and the stoichiometric uptake of PCB imply that the apoprotein autoassembled quantitatively. Furthermore, similar SAR values were obtained for preparations from clones coexpressing GroES and GroEL chaperonins. Such coexpression often has dramatic effects on the solubility, quarternary assembly and activity of recombinant proteins, including phytochromes, in E. coli (8, 33, 40). If the low SAR resulted from a proportion of the apoprotein that was unable to autoassemble or become spectrally active, differences would probably have emerged; we are, however, currently unable to exclude the possibility.
An alternative explanation for the low SAR of the recombinant phytochrome relates to the form of the chromophore in different molecular environments. We note that, whereas the Pr absorption peak at 658 nm is low, Soret absorption around 370 nm is high, relative to the situation in plant phytochromes. High Soret and low R absorbance is characteristic of helical tetrapyrroles whereas the opposite holds for extended tetrapyrroles (44–47). On this basis, the Synechocystis phytochrome–PCB spectrum can be interpreted as reflecting a more helical chromophore than is typical of plant phytochromes.
Recombinant Synechocystis phytochrome from our system is particularly pure, soluble, and easy to produce. The apoprotein folds appropriately and accumulates harmlessly to extraordinarily high levels in the cytosol of the E. coli host. The C-terminal histidine tag must be exposed and freely accessible to allow the efficient affinity purification we have demonstrated. The apoprotein is very soluble and attaches tetrapyrrole chromophore readily, giving photochromic phytochrome holoprotein. The data reported here and in the parallel work of Remberg et al. (6) imply close similarities between Synechocystis phytochrome and plant phytochromes in the N-terminal chromophore-bearing portion, providing a useful model for basic studies of phytochrome structure/function. This expression system represents a dramatic improvement over others known to us (20, 21, 35, 37, 39, 40, 48–52). Although we cannot rigorously exclude an effect of the affinity tag on the conformation of the molecule, the pF10.Stop product behaves indistinguishably from the tagged recombinant. Affinity-purified recombinant Synechocystis phytochrome is therefore an attractive material for crystallization and subsequent x-ray diffraction studies of phytochrome molecular structure.
Acknowledgments
We appreciate the contribution of the Kazusa Institute in making the Synechocystis genome data available to the scientific community. We are grateful to Prof. Kurt Schaffner (Max-Planck-Institut für Strahlenchemie, Mülheim) for generous support and Prof. Silvia Braslavski (Max-Planck-Institut für Strahlenchemie, Mülheim) for valuable discussions and suggestions. Crystalline phycocyanobilin was kindly provided by Ingo Lindner. We thank Sabine Buchert for excellent technical support. Part of this work was funded by the Deutsche Forschungsgemeinschaft (J.H., T.B., and T.L.).
ABBREVIATIONS
- AmS
ammonium sulfate
- FR
730 nm monochromatic far-red light
- PCB
phycocyanobilin
- PΦB
phytochromobilin
- R
658 nm monochromatic red light
- Pr
red-absorbing phytochrome
- Pfr
far-red-absorbing phytochrome
- SAR
specific absorbance ratio
Note Added in Proof
The recent report of Yeh et al. (53) demonstrates Pr-specific autophosphorylation of the holophytochrome and that D68 of the downstream response regulator is indeed a Pr-dependent phosphotransferase target.
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the following databases: CyanoBase [Synechocystis phytochrome (slr0473) and Synechocystis putative response regulator (slr0474)] and GenBank [Ceratodon phytochrome (U56698), E. coli RcsC osmoregulation sensory kinase (D28242), Synechocystis plpA phytochrome-like protein (U67397), and Fremyella RcaE chromatic adaptation sensor protein (U59741)].
References
- 1.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 2.Hughes J, Lamparter T, Mittmann F. Plant Physiol. 1996;112:446. [Google Scholar]
- 3.Hughes J, Lamparter T, Mittmann F, Hartmann E, Gärtner W, Wilde A, Börner T. Nature (London) 1997;386:663. doi: 10.1038/386663a0. [DOI] [PubMed] [Google Scholar]
- 4.Schneider-Poetsch H A W, Braun B, Marx S, Schaumburg A. FEBS Lett. 1991;281:245–249. doi: 10.1016/0014-5793(91)80403-p. [DOI] [PubMed] [Google Scholar]
- 5.Schneider-Poetsch H A W. Photochem Photobiol. 1992;56:839–846. doi: 10.1111/j.1751-1097.1992.tb02241.x. [DOI] [PubMed] [Google Scholar]
- 6.Remberg, A., Lindner, I., Lamparter, T., Hughes, J., Kneip, K., Hildebrand, P., Braslavsky, S. E., Gärtner, W. & Schaffner, K. (1997) Biochemistry, in press. [DOI] [PubMed]
- 7.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: A Laboratory Manual. NY: Cold Spring Harbor Lab. Press; Plainview; 1989. [Google Scholar]
- 8.Goloubinoff P, Gatenby A A, Lorimer G H. Nature (London) 1989;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- 9.Kufer W, Scheer H. Hoppe-Seylers Z Physiol Chem. 1979;360:935–941. doi: 10.1515/bchm2.1979.360.2.935. [DOI] [PubMed] [Google Scholar]
- 10.Cole W J, Chapman D J, Siegelman H W. J Am Chem Soc. 1967;89:3642–3645. [Google Scholar]
- 11.Cornejo J, Beale S I, Terry M J, Lagarias J C. J Biol Chem. 1992;267:14790–14798. [PubMed] [Google Scholar]
- 12.Bonfiglio J V, Bonnett R, Buckley D G, Hamzetash D, Hursthouse M B, Malik K M A, McDonagh A F, Trotter J. Tetrahedron. 1983;39:1865–1874. [Google Scholar]
- 13.Berkelman T R, Lagarias J C. Anal Biochem. 1986;156:194–201. doi: 10.1016/0003-2697(86)90173-9. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y-C, Stewart S J, Cordonnier M-M, Pratt L. Planta. 1991;184:96–104. doi: 10.1007/BF00208242. [DOI] [PubMed] [Google Scholar]
- 16.Butler W L, Hendricks S B, Siegelman H W. Photochem Photobiol. 1964;3:521–528. [Google Scholar]
- 17.Pratt L H. Photochem Photobiol. 1975;22:33–36. doi: 10.1111/j.1751-1097.1975.tb06717.x. [DOI] [PubMed] [Google Scholar]
- 18.Jayaratne P, Keenleyside W J, MacLachlan P R, Dodgson C, Whitfield C. J Bacteriol. 1993;175:5384–5394. doi: 10.1128/jb.175.17.5384-5394.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gärtner W, Hill C, Worm K, Braslavski S E, Schaffner K. Eur J Biochem. 1996;236:978–983. doi: 10.1111/j.1432-1033.1996.00978.x. [DOI] [PubMed] [Google Scholar]
- 21.Murphy J T, Lagarias J C. Photochem Photobiol. 1997;65:750–758. doi: 10.1111/j.1751-1097.1997.tb01920.x. [DOI] [PubMed] [Google Scholar]
- 22.Grimm R, Rüdiger W. Z Naturforsch C. 1986;41:988–992. [Google Scholar]
- 23.Kehoe D M, Grossman R. Science. 1996;273:1409–1412. doi: 10.1126/science.273.5280.1409. [DOI] [PubMed] [Google Scholar]
- 24.Wilde A, Churin Y, Schubert H, Börner T. FEBS Lett. 1997;406:89–92. doi: 10.1016/s0014-5793(97)00248-2. [DOI] [PubMed] [Google Scholar]
- 25.Hoch J A, Silhavy T J, editors. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol. Press; 1995. [Google Scholar]
- 26.Wurgler-Murphy S M, Saito H. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 27.Kakimoto T. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 28.Boylan M T, Quail P H. Protoplasma. 1996;195:12–17. [Google Scholar]
- 29.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Science. 1996;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 30.Lagarias D M, Wu S-H, Lagarias J C. Plant Mol Biol. 1995;29:1127–1142. doi: 10.1007/BF00020457. [DOI] [PubMed] [Google Scholar]
- 31.Levit M, Lui Y, Surette M, Stock J. J Biol Chem. 1996;271:32057–32063. doi: 10.1074/jbc.271.50.32057. [DOI] [PubMed] [Google Scholar]
- 32.Nifa E G, Atkinson M R, Kamberov E S, Nifa A J. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgerton M D, Santos M O, Jones A M. Plant Mol Biol. 1993;21:1191–1194. doi: 10.1007/BF00023616. [DOI] [PubMed] [Google Scholar]
- 34.Elich T D, McDonagh A F, Palma L A, Lagarias J C. J Biol Chem. 1989;264:183–189. [PubMed] [Google Scholar]
- 35.Li L, Lagarias J C. J Biol Chem. 1992;267:19204–19210. [PubMed] [Google Scholar]
- 36.Li L, Murphy J T, Lagarias J C. Biochemistry. 1995;34:7923–7930. doi: 10.1021/bi00024a017. [DOI] [PubMed] [Google Scholar]
- 37.Deforce L, Tomizawa K, Ito N, Farrens D, Song P-S, Furuya M. Proc Natl Acad Sci USA. 1991;88:10392–10396. doi: 10.1073/pnas.88.23.10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Lagarias J C. Proc Natl Acad Sci USA. 1994;91:12535–12539. doi: 10.1073/pnas.91.26.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunkel T, Tomizawa K, Kern R, Furuya M, Chua N-H, Schäfer E. Eur J Biochem. 1993;93:587–593. doi: 10.1111/j.1432-1033.1993.tb18069.x. [DOI] [PubMed] [Google Scholar]
- 40.Hill C, Gärtner W, Towner P, Braslavsky S E, Schaffner K. Eur J Biochem. 1994;223:69–77. doi: 10.1111/j.1432-1033.1994.tb18967.x. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt P, Westphal U H, Worm K, Braslavski S E, Gärtner W, Schaffner K. J Photochem Photobiol B. 1996;34:73–77. doi: 10.1016/1011-1344(95)07269-1. [DOI] [PubMed] [Google Scholar]
- 42.Kidd D G, Lagarias J C. J Biol Chem. 1990;12:7029–7035. [PubMed] [Google Scholar]
- 43.Lagarias J C, Kelly J M, Cyr K L, Smith W O. Photochem Photobiol. 1987;46:5–13. [Google Scholar]
- 44.Thümmler F, Rüdiger W. Tetrahedron. 1983;39:1943–1951. [Google Scholar]
- 45.Arciero D M, Bryant D A, Glazer A N. J Biol Chem. 1988;263:18343–18349. [PubMed] [Google Scholar]
- 46.Falk H. The Chemistry of Linear Oligopyrroles and Bile Pigments. Vienna: Springer; 1989. [Google Scholar]
- 47.Schirmer T, Bode W, Huber R, Sidler W, Zuber H. J Mol Biol. 1985;184:257–277. doi: 10.1016/0022-2836(85)90379-1. [DOI] [PubMed] [Google Scholar]
- 48.Wahleithner J A, Li L, Lagarias J C. Proc Natl Acad Sci USA. 1991;88:10387–10391. doi: 10.1073/pnas.88.23.10387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomizawa K, Ito N, Komeda Y, Uyeda T Q P, Takio K, Furuya M. Plant Cell Physiol. 1991;32:95–102. [Google Scholar]
- 50.Lamparter T, Hartmann E, Podlowski S, Schneider-Poetsch H A W, Hughes J. J Plant Physiol. 1995;147:426–434. [Google Scholar]
- 51.Kunkel T, Speth V, Bc̈he C, Schäfer E. J Biol Chem. 1996;270:20193–20220. doi: 10.1074/jbc.270.34.20193. [DOI] [PubMed] [Google Scholar]
- 52.Wu S H, Lagarias J C. Proc Natl Acad Sci USA. 1996;93:8989–8994. doi: 10.1073/pnas.93.17.8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeh K-C, Wu S-H, Murphy J T, Lagarias J C. Science. 1997;277:1515–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]