Abstract
Ligands usually bind to proteins by displacing water from the binding site. The affinity and kinetics of binding therefore depend on the hydration characteristics of the site. Here, we show that the extreme case of a completely dehydrated free binding site is realized for the large nonpolar binding cavity in bovine β-lactoglobulin. Because spatially delocalized water molecules may escape detection by x-ray diffraction, we use water 17O and 2H magnetic relaxation dispersion (MRD), 13C NMR spectroscopy, molecular dynamics simulations, and free energy calculations to establish the absence of water from the binding cavity. Whereas carbon nanotubes of the same diameter are filled by a hydrogen-bonded water chain, the MRD data show that the binding pore in the apo protein is either empty or contains water molecules with subnanosecond residence times. However, the latter possibility is ruled out by the computed hydration free energies, so we conclude that the 315 Å3 binding pore is completely empty. The apo protein is thus poised for efficient binding of fatty acids and other nonpolar ligands. The qualitatively different hydration of the β-lactoglobulin pore and carbon nanotubes is caused by subtle differences in water–wall interactions and water entropy.
Keywords: β-lactoglobulin, free energy simulation, hydrophobic hydration, magnetic relaxation dispersion
Globular proteins are stabilized by dense atomic packing and by water exclusion from the nonpolar core region. However, the packing density is not uniform and a typical protein contains approximately four cavities per 100 residues of sufficient size to accommodate at least one water molecule (1). Small nonpolar cavities are usually empty and can be regarded as packing defects, whereas small polar cavities tend to be occupied by structural water molecules that stabilize the folded protein by H-bonding to otherwise unsatisfied peptide partners (2). Large cavities are frequently linked to protein functions, such as ligand binding and transport, membrane translocation, and enzyme catalysis. Some of these large cavities are lined exclusively or predominantly by nonpolar side-chains. The question whether such large nonpolar cavities are empty or contain water molecules is of fundamental biological importance (3–8).
The principal tool of structural biology, x-ray crystallography, cannot easily resolve this issue. There are three potential problems. (i) Because water molecules interact only weakly with the nonpolar cavity walls, they tend to be positionally disordered. As a result of such delocalization, the electron density of the water molecules may fall below the detection limit. (ii) At a typical resolution limit of ≈2 Å, a contaminating nonpolar ligand may be misinterpreted as a water cluster (9, 10). (iii) For structures determined at cryogenic temperatures, the hydration status of the cavity may be altered by re-equilibration during the flash cooling process (11).
In favorable cases, water molecules in nonpolar protein cavities can be inferred from intermolecular nuclear Overhauser effects (NOEs) measured by solution NMR spectroscopy (12, 13). However, weak water–protein NOEs, as expected from disordered water molecules, are difficult to distinguish from intramolecular NOEs relayed by exchanging labile protons (14) or even from long-range NOEs to bulk water (15). Molecular dynamics and free-energy simulations can in principle provide detailed insights about cavity hydration, but their predictive power is limited by the known shortcomings of current force fields (16, 17).
Several attempts have been made to establish the hydration state of a nonpolar 40 Å3 cavity in human interleukin-1β. The two x-ray diffraction studies that focused on this issue came to different conclusions: the first study assigned two water molecules with 70% occupancy to the cavity (18), whereas the more recent one indicated that the cavity is empty (19). Free energy calculations on a nonpolar spherical model cavity indicated stable hydration by clusters of at least three cyclically H-bonded water molecules (20), but the interleukin-1β x-ray data are not consistent with a cluster of three or more water molecules unless the occupancy is very low (so the cavity is empty most of the time). In contradiction with the x-ray results, water NOEs with protein protons near the interleukin cavity were taken as evidence for two to four water molecules in the cavity (12) and a recent MD simulation study indicated stable hydration by a four-water cluster (21). The hydration status of the interleukin-1β cavity thus remains controversial.
Free-energy calculations on a large, weakly polar, globular cavity in the surface layer protein tetrabrachion predicted stable hydration by a cluster of seven to nine water molecules (22), in line with calculations on model cavities (20) and fully consistent with the crystal structure. Agreement between free energy calculations and x-ray crystallography was also found for a 160 Å3 nonpolar cavity engineered into T4 lysozyme, which was found to be empty under ambient conditions but cooperatively filled by several water molecules at kbar pressures (23).
Here, we investigate the hydration of a functional cavity that is both larger and less polar than any of the cavities mentioned above. Like other members of the lipocalin family, bovine β-lactoglobulin (BLG) has a β-barrel fold enclosing a cavity that binds nonpolar ligands, including palmitate (9, 24, 25). This cavity, known as the calyx, has a volume of 315 Å3 (as gauged by a 1.4 Å probe) with approximately cylindrical (5 × 15 Å) shape. The cavity is lined by 12 aliphatic and one aromatic side chain, with only one polar atom (Met-107.S) within 5 Å of the cavity axis. Access to the calyx is restricted by a flexible loop that forms a lid on the calyx entrance. Deprotonation of Glu-89, with an anomalously high pKa value, triggers a conformational transition that opens the lid (9, 26). Depending on ionic strength, this so-called Tanford transition occurs in the pH range 6–8.
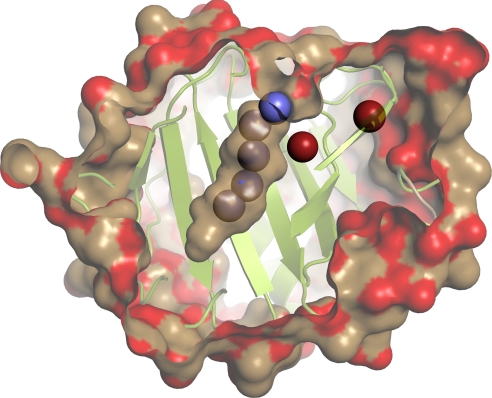
The available crystallographic evidence does not conclusively establish whether the calyx is hydrated in the apo form of BLG. In several structures with the loop in the open conformation (at pH > 7), no electron density was found in the calyx (9, 24). However, in the structure 3BLG, in the closed conformation at pH 6.2, the electron density observed in the calyx was modeled as a string of five water molecules (Fig. 1) within H-bonding distance (2.7–3.0 Å) of each other (9, 27). The C–C separations across the calyx at the positions of these five water molecules are 8 ± 1 Å, closely matching the corresponding dimension (8.1 Å) of the smallest carbon nanotube (CNT). Experimental and computational studies show that such CNTs are filled by a chain of water molecules under ambient conditions (28).
Fig. 1.
View of the crystal structure 3BLG (9) of β-lactoglobulin, showing two water molecules (red) buried in small polar cavities (labeled 175 and 177 in 3BLG, with 177 closer to the calyx) and five water molecules (blue) modeled in the nonpolar calyx. We label the calyx waters 1–5 starting from the bottom; the corresponding 3BLG labels are 178, 190, 179, 180, and 233. The cavity surface in the calyx is color-coded according to whether the contributing protein atoms are polar (red) or nonpolar (brown). The light green cartoon representation shows one of the two β sheets enclosing the nonpolar calyx. The figure was rendered with PyMOL (www.pymol.org) using a 1.4 Å probe to define the (external and internal) molecular surfaces of the protein and the same protein vdW radii as used for the cavity volume calculations.
To clarify the hydration status of the calyx, we have performed water 17O and 2H magnetic relaxation dispersion (MRD) measurements (29) on BLG in aqueous solution. Because palmitate displaces any water molecules in the calyx, we can directly monitor calyx hydration via the difference in the MRD data recorded on the apo protein and the palmitate–BLG complex. This difference-MRD approach has been used to characterize internal water molecules in polar cavities (30–32). The MRD data indicate that the calyx is devoid of long-lived (≥ ns) water, and the presence of more mobile water is ruled out by the free energy calculations reported here. We also obtain information about the smaller polar cavities, which are found to be occupied by ordered water molecules with residence times of ≈20 ns, consistent with our MD simulations.
Results
Identification of Internal Water Molecules.
Water 17O and 2H MRD profiles were recorded from aqueous solutions of BLG at neutral pH (Figs. 2 and 3). The apo form of BLG, without any hydrophobic ligand in the calyx, was examined at pH 7.4 (sample A1, calyx lid open) and at pH 6.2 (sample A2, calyx lid closed). Two independent samples (H1 and H2) of the holo form with a palmitate molecule bound in the calyx were studied, both at pH 7.4. At neutral pH, the protein exists predominantly as a homodimer. The dimer interface buries only 6% of the solvent-accessible surface area and does not interfere with ligand binding because the entrance to the calyx is 15 Å from the nearest dimer contact (9, 24). The structural changes in the protein induced by fatty acid binding are small and localized (25). Any observed difference between the concentration-normalized MRD profiles from the holo and apo proteins can therefore be attributed to water molecules in the calyx of the apo form, which are displaced by the bound palmitate molecule in the holo form. This argument assumes that the holo sample really has palmitate bound in the calyx. This was confirmed by 13C NMR spectroscopy, yielding 0.94 ± 0.03 (sample H1) and 0.91 ± 0.02 (sample H2) mole bound palmitate per mole BLG. If the calyx were hydrated as in Fig. 1, we would expect an MRD profile of substantially larger amplitude for the apo form than for the holo form. However, as seen from Figs. 2 and 3, the difference between the MRD profiles from the holo and apo forms is hardly significant. This observation places upper bounds on the number of water molecules in the calyx and on their order parameters and residence times (see Discussion).
Fig. 2.
Water 17O MRD profiles from aqueous solutions of β-lactoglobulin with (holo) or without (apo) bound palmitate at 27°C. The data symbols refer to apo BLG at pH 7.4 (open squares, sample A1) and pH 6.2 (open diamonds, sample A2); holo BLG at pH 7.4 (filled triangles, sample H1; inverted filled triangles, sample H2); and protein-free buffer (open circles). Data from different BLG solutions have been normalized to the protein concentration of sample A1 (0.79 mM, NW = 69300), using the scaling R1-R10 ∝ 1/NW implicit in Eq. 1. The curves were obtained by fitting the model to the two sets of apo or holo data as described in the text. The parameter values resulting from the fits are given in Table 1.
Fig. 3.
Water 2H MRD profiles from aqueous solutions of β-lactoglobulin with (holo) or without (apo) bound palmitate at 27°C. Symbols and curves as in Fig. 2. The 2H and 17O data were measured on the same samples.
The quantitative analysis of the MRD data is based on a model with two classes (α, β) of hydration water in fast exchange with bulk water. The β class contains the most long-lived internal water molecules, responsible for the dominant low-frequency component in the MRD profile. There are Nβ such water molecules, with mean-square orientational order parameter Sβ2. The α class comprises the remaining hydration waters, interacting with the protein surface. They have a wide distribution of correlation times, that may range from the bulk water correlation time, τ0 = 1.9 ps, up to the β-class correlation time, τβ = 11 ± 2 ns. We model this distribution with a power law, but this choice hardly affects the β-class parameters of primary interest here.
The curves in Figs. 2 and 3 were obtained by fitting three model parameters to the MRD data: the order-parameter-weighted occupancy (NβSβ2) and mean residence time (τW) of long-lived (>10−8 s) internal water molecules, and the power-law exponent (v) in the correlation time distribution for the external hydration layer. Because there is no significant difference between the two sets of data for the apo form (at pH 6.2 and 7.4), a single fit was performed on the combined apo data. Similarly, a single fit was performed on the combined data for the two equivalent holo samples. The parameter values resulting from these fits are collected in Table 1. The key finding is that there is no significant difference between the apo and holo samples in any of the three parameters. This is true for the 17O data as well as for the 2H data. Specifically, the 17O data yield NβSβ2(apo) − NβSβ2(holo) = −0.05 ± 0.13 with 95% confidence limits. This result rules out the presence of even a single moderately ordered water molecule in the calyx, provided that it has a residence time of order 10−8 s or longer.
Table 1.
Results of fits to water 17O and 2H MRD data
| Sample | Nuclide | NβSβ2 | τW, ns | v | χ2 |
|---|---|---|---|---|---|
| Apo (A1 and A2) | 17O | 2.6 ± 0.2 | 20 ± 2 | 1.91 ± 0.02 | 1.1 |
| Holo (H1 and H2) | 17O | 2.7 ± 0.2 | 18 ± 2 | 1.93 ± 0.02 | 0.7 |
| Apo (A1 and A2) | 2H | 3.2 ± 0.2 | 27 ± 3 | 1.93 ± 0.02 | 0.7 |
| Holo (H1 and H2) | 2H | 3.0 ± 0.2 | 28 ± 4 | 1.94 ± 0.02 | 0.5 |
The correlation time, τβ, deduced from MRD profiles of protein solutions usually matches the protein rotational correlation time, τRP, and can then only provide bounds on the water residence time, τW:τRP ≪ τW < 1/(ωQ2Sβ2τRP) ≈ 1 μs (29). Here, however, the observed correlation time τβ for both sets of 17O data are a factor 2 shorter than the rotational correlation time of the BLG dimer, τRP = 23 ns, allowing us to determine τW with the aid of Eq. 4. The long residence time, τW ≈ 20 ns, indicates that these water molecules are at least partly buried, but they cannot be located in the calyx because they are present in the apo form as well as in the holo form. The value NβSβ2(17O) = 2.6 ± 0.2 implies, because Sβ2 ≤ 1, that there are three or more water molecules with mean residence time τW ≈ 20 ns. Obvious candidates are the two water molecules (Fig. 1) that are buried in small polar cavities in nearly all crystal structures of BLG. In the structure 3BLG (9), water 175 makes three strong H-bonds to the protein backbone (Gln-35.O, Arg-40.O, Trp-61.N), and should therefore be highly ordered. Water 177 is deeply buried, but makes only one strong H-bond to the backbone (Leu-39.O) in addition to two weaker polar interactions with side-chains (Met-24.Sδ and Gln-120.Oε).
The 2H MRD profiles (Fig. 3) are similar to the 17O profiles (Fig. 2). As for 17O, there is no significant difference between the parameter values derived from the apo and holo profiles (Table 1). However, the 2H parameters NβSβ2 and τW are significantly larger than their 17O counterparts. These differences reflect a contribution from labile BLG hydrogens in hydroxyl, amino, and guanidino groups, exchanging with water deuterons on the millisecond time scale at pH 7.4. There are 82 such hydrogens in BLG, but they are in the slow-exchange regime and therefore add to NβSβ2 only a small fraction of their maximum contribution (29). The water contribution to NβSβ2(2H) is significantly less than NβSβ2(17O), because Sβ2(2H) < Sβ2 (17O) (see below). Therefore, as much as half of NβSβ2(2H) may be produced by labile hydrogens. Because these labile hydrogens have residence times in the millisecond range, much longer than τRP, the effective correlation time τβ should be longer for 2H than for 17O, as observed (Table 1). The 2H MRD data also rule out the (unlikely) possibility that the calyx contains water molecules with residence times in the range 1–100 μs, in which case they would contribute to the 2H MRD profile (fast exchange) but not to the 17O MRD profile (slow-exchange regime) (29).
Hydration Dynamics and Order.
To further characterize the hydration water, we performed molecular dynamics (MD) simulations of apo BLG in water at 300 K. The two internal water molecules buried in polar cavities (Fig. 1) exchanged once (W175) or twice (W177) during the 20-ns trajectory, consistent with the 20-ns mean residence time deduced from the MRD analysis. The orientational order parameter was also computed for these water molecules. The most strongly interacting of these, W175, is highly ordered with S2(17O) = 0.68 ± 0.07. The other crystallographically identified internal water molecule, W177, is less ordered. During most of the trajectory, it alternates between two nearby hydration sites. Averaging over both sites yields a relatively low order parameter, S2(17O) = 0.17 ± 0.07. However, during the last 4.5 ns of the trajectory, both sites are simultaneously occupied, with order parameters S2(17O) = 0.61 ± 0.05 (W177a) and 0.40 ± 0.05 (W177b). The sum of the S2 values for these three water molecules, 1.7 ± 0.1, is smaller than the experimental value, NβSβ2 = 2.6 ± 0.2. If the simulation yields accurate order parameters, there must be at least one additional long-lived, fully or partly buried, water molecule. Whereas the simulations were performed on monomeric BLG, 95% of the protein in the MRD sample is present in dimeric form [supporting information (SI) Text]. (The MRD parameter NβSβ2 is quoted on a monomer basis.) The difference between the experimental and computed NβSβ2 values might therefore be due to one or more long-lived water molecules at the dimer interface. In the BLG crystal dimer 1BSY (9), three water molecules are partly buried at the rim of the interface, each making two strong H-bonds with the protein.
Order parameters were also computed for the putative string of five water molecules in the calyx (Fig. 1). To prevent these water molecules from escaping from the calyx, they were harmonically restrained to their crystallographic positions with the same harmonic potential as used for the free energy calculations, but their orientations were not constrained. We thus obtained NβSβ2 = 1.3 ± 0.2 for the five water molecules in the calyx. Because NβSβ2(apo) − NβSβ2(holo) = −0.05 ± 0.13 with 95% confidence, we can rule out this hydration motif provided that the water molecules are sufficiently long-lived to contribute to the β dispersion (see Discussion).
The 2H order parameter is generally smaller than the 17O order parameter, because only the former is affected by 180° flips about the water C2 axis (29). If the C2 flip is fast compared with τβ (≈10 ns) and if other internal motions can be neglected, we expect Sβ2(17O)/Sβ2(2H) = 2.7 (29). The simulations show that this limit is reached for the weakest polar hydration site (W177b), whereas the ratio is 1.3 for W175 and W177a. Internal water molecules without H-bonds to the protein are likely to undergo fast flip motions and are therefore more strongly manifested in the 17O MRD profile. Indeed, for the five calyx sites, the computed ratio Sβ2(17O)/Sβ2(2H) is between 2.0 and 2.7.
From the power-law exponent, v = 1.92 ± 0.02 (Table 1), deduced from the 17O MRD profiles, we can obtain the product of the number, ΔNα, of surface hydration water molecules in a given correlation time range and their mean-square order parameter, Sα2. We thus find ΔNαSα2 = 12 ± 1 for τα > 100 ps. From the MD trajectory, we identify ΔNα = 17 ± 1 surface hydration sites that are occupied by water molecules with residence times longer than 100 ps. This is consistent with the experimental result: correlation times as long as 100 ps and above should reflect exchange out of the hydration site, rather than the much faster restricted rotation within the site which makes Sα2 < 1.
Hydration Thermodynamics.
To estimate the driving force for hydration of the nonpolar calyx, we used thermodynamic integration to compute the standard free energy change, ΔGhydo(n|single), for the process where a single water molecule is transferred from bulk water to one of the five crystallographic hydration sites (Fig. 1). We also computed the free energy change for the stepwise filling of the calyx with five water molecules, starting either from site 1 at the bottom, ΔGhydo(n|1 → 5), or from site 5 at the mouth, ΔGhydo(n|5 → 1). The free energies and the site occupancies, ξ(n), derived from them are collected in Table 2.
Table 2.
Hydration free energies and occupancies for calyx sites
| n* | ΔGhyd(n)†o |
10log ξ(n) |
||||
|---|---|---|---|---|---|---|
| Single | 1 → 5 | 5 → 1 | Single | 1 → 5 | 5 → 1 | |
| 1 | 24 ± 2 | 24 ± 2 | 12 ± 1 | −4.1 ± 0.4 | −4.2 ± 0.4 | −2.1 ± 0.2 |
| 2 | 23 ± 1 | 17 ± 2 | 14 ± 1 | −3.9 ± 0.1 | −2.9 ± 0.4 | −2.5 ± 0.2 |
| 3 | 12 ± 2 | 9 ± 2 | 19 ± 1 | −2.1 ± 0.4 | −1.6 ± 0.4 | −3.3 ± 0.2 |
| 4 | 14 ± 3 | 11 ± 4 | 7 ± 1 | −2.5 ± 0.6 | −1.8 ± 0.8 | −1.3 ± 0.2 |
| 5 | 10 ± 4 | 10 ± 2 | 7 ± 1 | −1.8 ± 0.6 | −1.8 ± 0.4 | −1.3 ± 0.2 |
| All | 71 ± 6 | 60 ± 3 | −12.4 ± 1.1 | −10.5 ± 0.4 | ||
*Hydration sites labeled as in Fig. 1.
†Free energies in kJ mol−1 refer to a standard concentration of 30.02Å −3 = 55.32 M.
All single-water site occupancies, ξ(n|single), are undetectably small (<1%), as expected because the nonpolar cavity cannot compensate for the loss of bulk water H-bonds. The free energy cost of filling all five hydration sites in the calyx is prohibitively large, 71 ± 6 kJ mol−1 (1 → 5) and 60 ± 3 kJ mol−1 (5 → 1). This corresponds to a probability of ≈10−11 for observing a fully hydrated calyx. The hydration motif depicted in Fig. 1 with a string of five water molecules in the calyx (9) is thus ruled out by the free-energy calculations.
Discussion
An observed difference between the 17O MRD profiles recorded before and after a well-defined molecular perturbation, such as palmitate binding to the calyx of BLG, would have provided unambiguous evidence for the presence of water in the calyx of the apo protein. Our finding that there is no significant difference does not categorically rule out the presence of water in the calyx, but it does impose bounds on the possible values of the order-parameter-weighted occupancy, NβSβ2, and on the mean residence time, τW, for any water molecules in the calyx. These bounds can be established by computing the difference MRD profile, ΔR1 = R1(apo) − R1(holo), for a range of NβSβ2 and τW values and comparing these with experimental ΔR1 data (Fig. S1). Given the experimental uncertainty, ΔR1 = 2 s−1, we can then divide the NβSβ2–τW plane into allowed and excluded regions (Fig. S2). Using the MD estimate, NβSβ2 = 1.3, we find that the MRD data rule out a five-water chain in the calyx if τW > 2 ns.
On the other hand, water molecules in the calyx would escape detection by MRD if they were sufficiently short-lived (τW < 2 ns). MD simulations of single-file water chains in CNTs yield residence times of order 0.1–1 ns (33). However, the calyx is closed at one end and is not as straight and smooth as a CNT, and these factors would slow down water exchange from the calyx. In particular, the flexible loop at the calyx mouth would partly block water exchange in the “closed” conformation at pH 6.2 (9, 26). Nevertheless, it is not certain that these factors would make τW > 2 ns. Therefore, we rely on the free energy calculations (Table 2) to rule out the case of short-lived calyx hydration. The methodological limitations in these calculations, notably the incomplete representation of polarization effects, are not likely to overturn the decisive outcome.
In conclusion, the experimental and theoretical results presented here are not consistent with a hydrated calyx in BLG. The electron density observed in the calyx in the crystal structure 3BLG must therefore be attributed to binding of a nonpolar impurity, a possibility that was entertained in the original publication (9) but that was later dismissed (27). Similar problems have been encountered with other lipocalins (10).
In the absence of other interactions than a hard confining potential, the mean water occupancy of a cavity (of arbitrary size and shape) is simply Vcav/Vvap, where Vcav is the (mean) volume of the (fluctuating) cavity and Vvap is the molar volume of water vapor in equilibrium with bulk water, which is 1.16 × 106Å3 at 300 K. Identifying Vcav with the volume, 27 Å3, accessible to the center of a 1.4 Å radius probe in the calyx of the 3BLG structure, divided by 6 because the calyx contains approximately six potential hydration sites, we obtain ξ(n|single) = 4 × 10−6. As expected, the computed single-water occupancies are larger, >10−4 (Table 2), indicating that water–protein interactions provide significant stabilization. Direct calculation of the mean water–protein interaction energy from the 10-ns MD simulation with “forced” hydration of the calyx yields −16 ± 3 kJ mol−1 for each of the five hydration sites (Table S1), essentially the same value as found for a spontaneously hydrated CNT of similar dimensions (28).
Polarization effects are likely to contribute to the different (observed) hydration of CNT and calyx. In contrast to the CNT, with polarizable π electrons in the graphene sheet, the calyx is lined by 12 aliphatic (Leu-39, Val-41, Val-43, Leu-46, Leu-54, Ile-56, Leu-58, Ile-71, Ile-84, Leu-87, Val-92, Leu-103) and only one aromatic (Phe-105) side chain. The incomplete treatment of polarization effects in the force field used in both of these simulations is likely to underestimate the stabilization energy in the CNT (because the O H···π interaction is not properly described by the Lennard–Jones parameters) and overestimate it in the calyx (because the TIP3P effective dipole moment is larger than expected for a single water molecule in a nonpolar environment).
H···π interaction is not properly described by the Lennard–Jones parameters) and overestimate it in the calyx (because the TIP3P effective dipole moment is larger than expected for a single water molecule in a nonpolar environment).
Hydration of nonpolar cavities can be promoted by electric fields produced by remote charges and dipoles (12, 34). A simple model calculation, taking into account the dipole moment (μ = 1.855 D) and polarizability (α = 1.662 × 10−40 C m2 V−1) of the water molecule in a singly occupied cavity, shows that an electric field E enhances the occupancy ξ above the value ξ0 for an ideal cavity according to ξ/(1− ξ) = ξ0 exp(A) sinh(M)/M, where A = α E2/(2kBT) and M = μE/(kBT). It thus follows that a field of ≈10 GV m−1 is required to obtain roughly half occupancy of a small nonpolar cavity, whereas a field of <5 GV m−1 has negligible effect (ξ < 10−3 in our case). We computed the protein-generated electric field E at the oxygen of internal water molecules from the 10-ns MD trajectory, finding 12.5 GV m−1 for W175, in the typical range for small polar cavities, but only 1.7–2.8 GV m−1 for the three deepest calyx sites (Table S1). Calyx hydration should thus not be significantly enhanced by protein-induced water polarization. Entropic factors also make the calyx more hydrophobic than a CNT. By occupying space, water molecules in the calyx suppress conformational fluctuations of the lining nonpolar side-chains, thereby increasing the free energy.
The total interaction energy per water molecule in the five-water chain (Fig. 1) is about −60 kJ mol−1 (Table S1), ≈15% less than in the CNT (28). Each water–water interaction within the calyx contributes about −20 kJ mol−1, as expected for a well formed H-bond with mean O–O separation of 2.84 Å (Table S1). Each water molecule in the chain switches H-bond donor and rotates about the calyx axis on a picosecond time scale, consistent with the relatively small order parameters and the large Sβ2(17O)/Sβ2(2H) ratios. Yet, the H-bonds are highly linear, with mean (OH,OO) angle of 15–25°. Higher water ordering (lower entropy) may thus contribute, along with the slightly weaker stabilization energy, to explain why the calyx is dehydrated while the CNT is hydrated.
BLG belongs to the calycin superfamily (35). The calycins share a common antiparallel β-barrel folding motif, and they all bind nonpolar molecules in the enclosed cavity. However, the mechanism and energetics of ligand binding differ widely among the calycins. For example, rat intestinal fatty acid binding protein (I-FABP), an abundant cytoplasmic calycin, binds palmitate (in an extended conformation, as in BLG) in a ≈500 Å3 globular cavity of mixed polarity and occupied by 20–30 water molecules (31, 36). This contrasts sharply with the completely nonpolar, empty binding cavity in BLG that seems to be poised for receiving an acyl chain or other nonpolar ligand. Yet, the palmitate affinity is lower for BLG (≈106 M−1) than for I-FABP (≈108 M−1) (37, 38). The different strategies for fatty acid binding used by these proteins may provide important clues to their still incompletely understood biological functions. All but one of the 13 nonpolar side-chains lining the calyx in BLG are conserved, or replaced by another nonpolar residue, in human glycodelin and in mouse major urinary protein (35). The binding cavity in the latter protein is not entirely nonpolar and appears to contain ≈4 water molecules that are displaced by the pheromone ligand (6). A recent MD simulation indicates that the binding cavity of apo cyclooxygenase-2 is empty 80% of the time (8), but the crystal structure of the complex with arachidonic acid shows 26 polar atoms, including five water molecules, within 5 Å of the ligand (39). To the best of our knowledge, BLG is the only clearly documented case of a large dehydrated binding cavity.
What are the biological implications of the complete exclusion of water from the 315 Å3 calyx in BLG? It has been suggested that a hydrophobic pore in a membrane protein may act as a “vapor lock,” where polarity-modulating conformational changes control the permeability to polar molecules by expelling water molecules in a microscopic version of capillary evaporation (or dewetting) (4, 40). In a hydrophobic pore with only one entrance, the effect of the void is instead to facilitate binding of nonpolar ligands. In I-FABP, the water molecules that are displaced as the ligand enters the cavity can exit through another transient channel. Because such a “back door” does not exist in BLG, productive ligand binding can only occur when the calyx has been evacuated. Stable hydration of the calyx would therefore drastically reduce the rate of ligand association as well as the binding affinity.
Materials and Methods
Sample Preparation and Characterization.
Bovine β-lactoglobulin isoform A was purified and complexed with 13C-labeled palmitate as detailed in SI Text. The amount of palmitate bound to BLG in the holo samples was determined by 13C NMR spectroscopy as described in SI Text.
Magnetic Relaxation Dispersion.
The longitudinal relaxation rate, R1, of the water 2H and 17O magnetizations was measured with 0.5–1.0% accuracy at 27.0 ± 0.1°C using five different NMR spectrometers, as described in SI Text. Fits to the MRD data were performed with the Levenberg–Marquardt algorithm and the parameter errors quoted in Table 1 were obtained with the Monte Carlo method using 1,000 synthetic data sets.
The MRD data were interpreted in terms of a model with two classes of hydration water (α, β) in fast exchange (on the spin relaxation time scale) with bulk water. The observed spin relaxation rate can then be decomposed as (29)
 |
where R10 is the relaxation rate measured on the protein-free buffer solution (open circles in Figs. 2 and 3) and the angular brackets signify an average over all water molecules in the class. NW is the analytically determined water/BLG mole ratio; see SI Text. Class α comprises the Nα (≈500) water molecules in contact with the external protein surface, whereas class β contains a small number of internal water molecules with mean residence time τW comparable to, or longer than, the rotational correlation time τRP (23 ns) of the dimeric protein. The average intrinsic relaxation rates in Eq. 1 are given by (29)
and similarly for 〈R1α(ω0)〉. Here, ωQ is the known nuclear quadrupole frequency (29) and the motional spectral density for class β has the Lorentzian form
where Sβ is the root-mean-square orientational order parameter (29) and the correlation time τβ is determined by protein tumbling and/or internal water exchange according to:
For further details, see SI Text.
Molecular Dynamics Simulations.
MD simulations of monomeric BLG in water were performed with the NAMD code (41) and the AMBER ff99 force field (42). The protein, with atomic coordinates from the crystal structure 3BLG (9) and with added H atoms and disulfide bonds, was solvated with 6,249 TIP3P water molecules (including the crystal waters) in a truncated octahedron. The net charge of the protein, with ionizable residues modeled in their dominant protonation states at neutral pH (also Glu-89 was deprotonated), was neutralized by nine sodium ions. After energy minimization and equilibration, the MD simulation was performed at 300 K and 1 atm with periodic boundary conditions, particle-mesh Ewald summation for long-range electrostatics, 10 Å cutoff for nonbonded interactions, constrained X-H bonds, and 2-fs time step. Atomic coordinates were saved every 2 ps of the 20-ns MD trajectory. Order parameters, electric fields and mean interaction energies for internal water molecules were calculated as described in SI Text.
Free Energy Calculations.
The standard free energy of hydration, ΔGhydo(n), for the process where a water molecule is transferred from bulk water to hydration site n in the calyx (labeled as in Fig. 1) was computed by thermodynamic integration using the double-decoupling method (43, 44) as described in SI Text. Calculations were performed for hydration of the calyx by a single water molecule, ΔGhydo(n), and for sequential addition of water molecules to all of the five hydration sites shown in Fig. 1. The latter filling process was carried out in two ways; starting from the bottom, ΔGhydo(n|1 → 5), or the mouth, ΔGhydo(n|5 → 1), of the calyx. With the standard state concentration corresponding to bulk water at 300 K (55.32 M), the site occupancy ξ(n) is related to the standard free energy of hydration as
Supplementary Material
Acknowledgments.
We thank Hanna Nilsson for protein purification, Torbjörn Drakenberg for help with 13C NMR experiments, Hans Lilja for NMR spectrometer maintenance, Ulf Ryde for help with simulations in Lund, Andrew McCammon for providing facilities for the initial computations, and Gerhard Hummer for valuable comments. This work was supported by the Swedish Research Council. Computational resources were financed by National Science Foundation and Howard Hughes Medical Institute (at University of California, San Diego) and by The Lund Centre for Computational Science (at Lund University).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709844105/DCSupplemental.
References
- 1.Rother K, Preissner R, Goede A, Frömmel C. Inhomogeneous molecular density: reference packing densities and distribution of cavities within proteins. Bioinformatics. 2003;19:2112–2121. doi: 10.1093/bioinformatics/btg292. [DOI] [PubMed] [Google Scholar]
- 2.Park S, Saven JG. Statistical and molecular dynamics studies of buried waters in globular proteins. Proteins. 2005;60:450–463. doi: 10.1002/prot.20511. [DOI] [PubMed] [Google Scholar]
- 3.Liong EC, Dou Y, Scott EE, Olson JS, Phillips GN. Waterproofing the heme pocket. J Biol Chem. 2001;276:9093–9100. doi: 10.1074/jbc.M008593200. [DOI] [PubMed] [Google Scholar]
- 4.Anishkin A, Sukharev S. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys J. 2004;86:2883–2895. doi: 10.1016/S0006-3495(04)74340-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu P, Huang X, Zhou R, Berne BJ. Observation of a dewetting transition in the collapse of the melittin tetramer. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 6.Barratt E, Bingham RJ, Warner DJ, Laughton CA, Phillips SEV, Homans SW. Van der Waals interactions dominate ligand-protein association in a protein binding site occluded from solvent water. J Am Chem Soc. 2005;127:11827–11834. doi: 10.1021/ja0527525. [DOI] [PubMed] [Google Scholar]
- 7.Tashiro M, Stuchebrukhov AA. Thermodynamic properties of internal water molecules in the hydrophobic cavity around the catalytic center of cytochrome c oxidase. J Phys Chem B. 2005;109:1015–1022. doi: 10.1021/jp0462456. [DOI] [PubMed] [Google Scholar]
- 8.Young T, Abel R, Kim B, Berne BJ, Friesner RA. Motifs for molecular recognition exploiting hydrophobic enclosure in protein-ligand binding. Proc Natl Acad Sci USA. 2007;104:808–813. doi: 10.1073/pnas.0610202104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin BY, Bewley MC, Creamer LK, Baker HM, Baker EN, Jameson GB. Structural basis of the Tanford transition of bovine β-lactoglobulin. Biochemistry. 1998;37:14014–14023. doi: 10.1021/bi981016t. [DOI] [PubMed] [Google Scholar]
- 10.Kuser PR, Franzoni L, Ferrari E, Spisni A, Polikarpov I. The X-ray structure of a recombinant major urinary protein at 1.75 Å resolution. A comparative study of X-ray and NMR-derived structures. Acta Crystallogr D. 2001;57:1863–1869. doi: 10.1107/s090744490101825x. [DOI] [PubMed] [Google Scholar]
- 11.Halle B. Biomolecular cryocrystallography: Structural changes during flash-cooling. Proc Natl Acad Sci USA. 2004;101:4793–4798. doi: 10.1073/pnas.0308315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst JA, Clubb RT, Zhou H-X, Gronenborn AM, Clore GM. Demonstration of positionally disordered water within a protein hydrophobic cavity by NMR. Science. 1995;267:1813–1817. doi: 10.1126/science.7892604. [DOI] [PubMed] [Google Scholar]
- 13.Otting G, Liepinsh E, Halle B, Frey U. NMR identification of hydrophobic cavities with low water occupancies in protein structures using small gas molecules. Nat Struct Biol. 1997;4:396–404. doi: 10.1038/nsb0597-396. [DOI] [PubMed] [Google Scholar]
- 14.Otting G. NMR studies of water bound to biological molecules. Progr NMR Spectrosc. 1997;31:259–285. [Google Scholar]
- 15.Halle B. Cross-relaxation between macromolecular and solvent spins: The role of long-range dipole couplings. J Chem Phys. 2003;119:12372–12385. [Google Scholar]
- 16.Jorgensen WL, McDonald NA, Selmi M, Rablen PR. Importance of polarization for dipolar solutes in low-dielectric media: 1,2-dichloroethane and water in cyklohexane. J Am Chem Soc. 1995;117:11809–11810. [Google Scholar]
- 17.Olano LR, Rick SW. Hydration free energies and entropies for water in protein interiors. J Am Chem Soc. 2004;126:7991–8000. doi: 10.1021/ja049701c. [DOI] [PubMed] [Google Scholar]
- 18.Yu B, Blaber M, Gronenborn AM, Clore GM, Caspar DLD. Disordered water within a hydrophobic protein cavity visualized by x-ray crystallography. Proc Natl Acad Sci USA. 1999;96:103–108. doi: 10.1073/pnas.96.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quillin ML, Wingfield PT, Matthews BW. Determination of solvent content in cavities in IL-1β using experimentally phased electron density. Proc Natl Acad Sci USA. 2006;103:19749–19753. doi: 10.1073/pnas.0609442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaitheeswaran S, Yin H, Rasaiah JC, Hummer G. Water clusters in nonpolar cavities. Proc Natl Acad Sci USA. 2004;101:17002–17005. doi: 10.1073/pnas.0407968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somani S, Chng C-P, Verma CS. Hydration of a hydrophobic cavity and its functional role: A simulation study of human interleukin-1β. Proteins. 2007;67:868–885. doi: 10.1002/prot.21320. [DOI] [PubMed] [Google Scholar]
- 22.Yin H, Hummer G, Rasaiah JC. Metastable water clusters in the nonpolar cavities of the thermostable protein tetrabrachion. J Am Chem Soc. 2007;129:7369–7377. doi: 10.1021/ja070456h. [DOI] [PubMed] [Google Scholar]
- 23.Collins MD, Hummer G, Quillin ML, Matthews BW, Gruner SM. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc Natl Acad Sci USA. 2005;102:16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brownlow S, Cabral JHM, Cooper R, Flower DR, Yewdall SJ, Polikarpov I, North ACT, Sawyer L. Bovine β-lactoglobulin at 1.8 Å resolution—still an enigmatic lipocalin. Structure. 1997;5:481–495. doi: 10.1016/s0969-2126(97)00205-0. [DOI] [PubMed] [Google Scholar]
- 25.Wu S-Y, Pérez MD, Puyol P, Sawyer L. β-Lactoglobulin binds palmitate within its central cavity. J Biol Chem. 1999;274:170–174. doi: 10.1074/jbc.274.1.170. [DOI] [PubMed] [Google Scholar]
- 26.Ragona L, Fogolari F, Catalano M, Ugolini R, Zetta L, Molinari H. EF loop conformational change triggers ligand binding in β-lactoglobulins. J Biol Chem. 2003;278:38840–38846. doi: 10.1074/jbc.M306269200. [DOI] [PubMed] [Google Scholar]
- 27.Jameson GB, Adams JJ, Creamer LK. Flexibility, functionality and hydrophobicity of bovine β-lactoglobulin. Int Dairy J. 2002;12:319–329. [Google Scholar]
- 28.Hummer G, Rasaiah JC, Noworyta JP. Water conduction through the hydrophobic channel of a carbon nanotube. Nature. 2001;414:188–190. doi: 10.1038/35102535. [DOI] [PubMed] [Google Scholar]
- 29.Halle B, Denisov VP, Venu K. Multinuclear relaxation dispersion studies of protein hydration. In: Krishna NR, Berliner LJ, editors. Biological Magnetic Resonance. New York: Kluwer Academic/Plenum; 1999. pp. 419–484. [Google Scholar]
- 30.Denisov VP, Peters J, Hörlein HD, Halle B. Using buried water molecules to explore the energy landscape of proteins. Nat Struct Biol. 1996;3:505–509. doi: 10.1038/nsb0696-505. [DOI] [PubMed] [Google Scholar]
- 31.Modig K, Rademacher M, Lücke C, Halle B. Water dynamics in the large cavity of three lipid-binding proteins monitored by17O magnetic relaxation dispersion. J Mol Biol. 2003;332:965–977. doi: 10.1016/s0022-2836(03)00968-9. [DOI] [PubMed] [Google Scholar]
- 32.Denisov VP, Schlessman JL, García-Moreno E. B., Halle B. Stabilization of internal charges in a protein: Water penetration or conformational change? Biophys J. 2004;87:3982–3994. doi: 10.1529/biophysj.104.048454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waghe A, Rasaiah JC, Hummer G. Filling and emptying kinetics of carbon nanotubes in water. J Chem Phys. 2002;117:10789–10795. [Google Scholar]
- 34.Vaitheeswaran S, Rasaiah JC, Hummer G. Electric field and temperature effects on water in the narrow nonpolar pores of carbon nanotubes. J Chem Phys. 2004;121:7955–7965. doi: 10.1063/1.1796271. [DOI] [PubMed] [Google Scholar]
- 35.Åkerström B, Flower DR, Salier J-P. Lipocalins: Unity in diversity. Biochim Biophys Acta. 2000;1482:1–8. doi: 10.1016/s0167-4838(00)00137-0. [DOI] [PubMed] [Google Scholar]
- 36.Sacchettini JC, Gordon JI. Rat intestinal fatty acid binding protein. J Biol Chem. 1993;268:18399–18402. [PubMed] [Google Scholar]
- 37.Kontopidis G, Holt C, Sawyer L. The ligand-binding site of bovine β-lactoglobulin: Evidence for a function? J Mol Biol. 2002;318:1043–1055. doi: 10.1016/S0022-2836(02)00017-7. [DOI] [PubMed] [Google Scholar]
- 38.Richieri GV, Ogata RT, Kleinfeld AM. Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J Biol Chem. 1994;269:23918–23930. [PubMed] [Google Scholar]
- 39.Kiefer JR, Pawlitz JL, Moreland KT, Stegeman RA, Hood WF, Gierse JK, Stevens AM, Goodwin DC, Rowlinson SW, Marnett LJ, et al. Structural insights into the stereochemistry of the cyclooxygenase reaction. Nature. 2000;405:97–101. doi: 10.1038/35011103. [DOI] [PubMed] [Google Scholar]
- 40.Beckstein O, Sansom MSP. Liquid-vapor oscillations of water in hydrophobic nanopores. Proc Natl Acad Sci USA. 2003;100:7063–7068. doi: 10.1073/pnas.1136844100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 43.Gilson MK, Given JA, Bush BL, McCammon JA. The statistical-thermodynamic basis for computation of binding affinities: A critical review. Biophys J. 1997;72:1047–1069. doi: 10.1016/S0006-3495(97)78756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamelberg D, McCammon JA. Standard free energy of releasing a localized water molecule from the binding pockets of proteins: Double-decoupling method. J Am Chem Soc. 2004;126:7683–7689. doi: 10.1021/ja0377908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.