Abstract
Most of the mathematical models that were developed to study the UK 2001 foot-and-mouth disease epidemic assumed that the infectiousness of infected premises was constant over their infectious periods. However, there is some controversy over whether this assumption is appropriate. Uncertainty about which farm infected which in 2001 means that the only method to determine if there were trends in farm infectiousness is the fitting of mechanistic mathematical models to the epidemic data. The parameter values that are estimated using this technique, however, may be influenced by missing and inaccurate data. In particular to the UK 2001 epidemic, this includes unreported infectives, inaccurate farm infection dates and unknown farm latent periods. Here, we show that such data degradation prevents successful determination of trends in farm infectiousness.
Keywords: foot-and-mouth disease, mathematical model, epidemiology
1. Introduction
In 2001, the UK experienced a major epidemic of foot-and-mouth disease (Anderson 2002). In all, 2026 premises across the country reported infected animals. In order to eliminate the disease from the UK, animals were culled on infected premises (IPs), and pre-emptively culled on farms thought to be at high risk of harbouring the disease or of becoming infected. One of the most controversial culling strategies was that of animals on farms contiguous to IPs. Out of the 8131 pre-emptively culled premises, 3369 were designated contiguous premises (CPs).
Mathematical models developed during and after the epidemic have shown that pre-emptive culling was effective in bringing the epidemic under control (Ferguson et al. 2001a,b; Kao 2001, 2003; Keeling et al. 2001; Ap Dewi 2004). However, a key assumption of all the models was that the infectiousness of IPs was constant over their infectious periods. This has lead to considerable controversy because if IP infectiousness increases during infectious period, CP culling may have been less effective at controlling the epidemic than has been suggested (Ferguson et al. 2001a).
Ferguson et al. (2001a) looked at how variable infectiousness altered the epidemic and the consequent control measures required to eliminate the disease. They included a parameter that allowed for a change in IP infectiousness after reporting. When infectiousness after reporting was equal to that before reporting, control measures greater than just rapid IP slaughter (e.g. CP pre-emptive culling) were predicted to be necessary to bring the epidemic under control. However, if infectiousness after reporting was five times higher than before reporting, rapid slaughter of IPs alone was sufficient to control the epidemic.
What determines the infectiousness of a farm is poorly understood. Some analyses suggest that animal species and numbers are important determinants (Keeling et al. 2001). It is probably true that farm biosecurity is another important determinant for both infectiousness and susceptibility to infection. However, quantifying biosecurity and its effect on transmission is extremely difficult. However, it is unknown if time-dependent processes change infectiousness. These include, for example, the amount of virus excreted by animals, which increases by several orders of magnitude during the first few days of infection (Hyslop 1965), the number of infectious animals on a farm, and farmers' responses to reported infections in their local area and on their farms.
The only practical method of estimating trends in infectiousness for the UK 2001 epidemic is the fitting of mechanistic mathematical models to the epidemic data. However, estimated parameter values from model fitting have to be tested for their sensitivity or robustness to the quality of the data the model is being fitted to. There are several such issues with the UK 2001 epidemic data. First, we do not know, in most cases, which farm infected which; tracing of transmission is notoriously difficult especially between farms where there are many possible routes of infection. Second, infection dates and latent periods are only estimates. Both are educated guesses based on tracing, age of lesions and veterinary experience. Third, we are not sure if all IPs were infected. Only 1340 (66%) IPs were tested positive for infection (Ferris et al. 2006), although a negative or inconclusive result does not necessarily imply that a farm was not infected. Finally, it is highly probable that not all infected farms were reported as infected because they were pre-emptively culled before clinical signs appeared. Such missing and inaccurate data could bias parameter estimates and reduce their significance. Thus, statistical tests may not be able to show that trends in infectiousness exist even if they do.
To date, there is no evidence for or against trends in infectiousness in the UK 2001 epidemic. Our aim in this paper is to determine if the available data collected during the epidemic are of sufficient quality to test the hypothesis that infectiousness varied over infectious period.
We will do this by fitting a mathematical model to the data. The model is closely based on another model that was developed during the 2001 epidemic and further used to study the vaccination control strategies in future outbreaks (Keeling et al. 2001; Tildesley et al. 2006). Our new model allows for changing infectiousness over infectious period. The model is fitted to the UK 2001 epidemic using maximum likelihood, and parameter estimates and their confidence intervals (CI) are determined. The sensitivity of parameter significance is analysed using simulated epidemics with the different types of missing and inaccurate data.
2. Methods
2.1 Demographic data
Ideally, we require the whole UK livestock demography in February 2001; unfortunately, this will never be known. One predictor of this demography is the June 2000 agricultural census of livestock premises combined with the Department of the Environment and Rural Affairs (DEFRA) list of livestock premises (approx. 186 000 farms). This dataset has been extensively analysed and cleaned and is the one we use here. Clearly, though, there are several problems in using this data. First and foremost are the numerous animal births, deaths and movements that occurred between June and February and beyond. Second, about 43 000 livestock premises are not recorded in the census that are in the DEFRA list of premises. These farms are recorded as having livestock present but with no indication of animal numbers. Finally, the recording of census information is not always accurate, especially for sheep in England and Wales (P. Bessell 2006, personal communication).
The data recorded in the epidemic Disease Control System has more accurate information on animal numbers culled; obviously, this information is only available for farms that had animals culled on them. The only animal numbers we have for the remaining farms are from the census, and an understanding of disease transmission requires knowledge of these farms as well. The Disease Control System data have, therefore, been used to validate the census data (P. Bessell 2006, personal communication). A consequence of using the June 2000 census is that the parameter estimates refer to the demography as recorded in the census and not to the true demography in February 2001.
2.2 UK 2001 epidemic data
The Disease Control System data contain information on all farms that were infected or pre-emptively culled. Of particular interest to this work are the estimated infection dates, the reporting dates and the cull dates. Reporting and cull dates are assumed to be accurate. Infection dates, however, were usually estimated by dating the age of lesions in animals (Gibbens et al. 2001) with additional assumptions about latent period and possible windows of transmission from other farms (Anderson 2002). They are, therefore, liable to considerable error.
The database also includes possible tracings between farms. In theory, this additional information could be used to inform the fitting of models. In reality, the tracings are scarce and their quality is poor, particularly local spread after the nationwide movement ban on 23 February 2001. Therefore, this information is not included in our analyses.
2.3 Assumptions about IP latent periods in 2001
IP latent periods are needed to determine infectious periods. Unfortunately, these are not known; previous modelling studies have assumed periods from 1 to 5 days (Ferguson et al. 2001a,b; Keeling et al. 2001; Tildesley et al. 2006). We assign IP latent periods as follows. IPs are first split into those in which infection was reported on or before being slaughtered and those in which infection was reported after being slaughtered. We assume that reporting before slaughter implies that animals on an IP passed through their latent periods were showing clinical symptoms and were therefore infectious. Out of the 2026 IPs, 1959 were reported on or before slaughter. The remaining 67, mainly dangerous contacts (DCs) later found to be infected, were reported after slaughter; either these IPs were never infectious, or they were and clinical signs were not present or were never spotted.
For IPs on which infection was reported on or before slaughter, we assume that their latent periods each last l days. Fixing the value of l, however, means that some IPs will be reported before becoming infectious; for these IPs (of which there are only 11 for l=5 days), we assume that their latent periods are equal to the time from infection to being reported. For IPs slaughtered before reporting, we again assume a latent period of l days. If an IP was slaughtered before the latent period ended, we assume that it was never infectious (only six cases for l=5 days). If we let td be the day an IP became infected, ts the day it became infectious, tr the day it was reported and tc the day it was slaughtered, then for IPs reported on or before slaughter we have
| (2.1) |
and for IPs reported after slaughter
| (2.2) |
2.4 The model
The model is based on a farm-level spatial model of the UK 2001 epidemic that includes farm heterogeneity (Keeling et al. 2001). The probability of a susceptible farm i being infected on day t is given by
| (2.3) |
where i is the susceptibility of farm i given by
| (2.4) |
j is the transmissibility of IP j given by
| (2.5) |
where Scattle is cattle susceptibility (sheep are assumed to have a susceptibility of 1); Tsheep and Tcattle are, respectively, sheep and cattle transmissibilities; Nsheep,j and Ncattle,j are sheep and cattle numbers on farm j; and ts,j is the day IP j becomes infectious. The function W(t−ts,j) describes change in infectiousness over infectious period. We assume that infectiousness is linearly related to time since becoming infectious and becomes constant once disease has been reported on the farm. Thus
| (2.6) |
where ρ is the rate of change of infectiousness from the first day of being infectious up to the report date and μ is the proportional change in infectiousness from report date onwards compared to the first day of being infectious. This reflects the potential change in infectiousness after disease has been reported. Both ρ and μ are assumed constant across all farms. Farms are considered non-infectious the day after being culled. Other forms for W(t) could be used, in particular an exponentially increasing infectiousness. The advantage of the linear form, however, is not only its simplicity, but also that it will fit an increasing trend in infectiousness whatever the form of that trend. If a trend is found using the linear form, then further analysis can be done to determine the precise form of the trend.
K(di,j) is the spatial transmission kernel given by
| (2.7) |
This is a slight modification from previous work (Keeling et al. 2001), incorporating a constant risk of infection over the first δ km, thus modelling spread between very close (e.g. contiguous) farms.
2.5 Statistical analysis
Before the nationwide movement ban on 23 February 2001, infection was generally, but not wholly, long range owing to the nationwide movement of infected sheep. After the ban, infection generally became localized to nearby farms. Thus, the epidemics seeded in the various regions of the UK became decoupled. We can therefore treat the regional epidemics separately after 23 February. We apply the model to seven regional epidemics; in order of size they are: North Cumbria, Welsh borders, South Cumbria, Dumfries and Galloway, Devon, Settle and Durham. Farms included in a region are specified by spatial and temporal windows defined in table 1.
Table 1.
Definition of regions on which analyses were performed.
| region | spatial window | temporal window | IPs | pre-emptive culls |
|---|---|---|---|---|
| North Cumbria | Cumbria above Northing 525 000 | 24 Feb–30 Jul | 672 | 2140 |
| Welsh borders | Wales, Shropshire, Gloucestershire, Hereford and Worcester and Avon | 24 Feb–19 Aug | 269 | 1389 |
| South Cumbria | Cumbria below Northing 525 000 | 11 May–30 Sep | 220 | 811 |
| Dumfries and Galloway | Dumfries and Galloway | 24 Feb–31 May | 176 | 1148 |
| Devon | Devon | 24 Feb–20 Jun | 102 | 435 |
| Settle | Lancashire and North Yorkshire west of Easting 437 000 | 25 Apr–24 Aug | 171 | 774 |
| Durham | Durham and Cleveland | 24 Feb–10 Jun | 96 | 288 |
The model is fitted to the UK 2001 epidemic data and simulated epidemic data using maximum likelihood. We assume that only IPs were infected in the UK 2001 epidemic, not CPs and DCs. For the simulated epidemics, we relax this assumption to determine its effect on estimated parameter values.
The log-likelihood function is given by (Keeling et al. 2001)
| (2.8) |
The maximum log-likelihood lmax for each region is found using the downhill simplex method (Press et al. 1992). The maximum-likelihood estimates of all parameters are found although, here, we report only values for ρ and μ which describe changes in infectiousness. The other fitted parameters were checked for their consistency to their true values. The 95% CIs on the parameter estimates are found using the profile likelihood. This is done by fixing the parameter of interest and maximizing equation (2.8) on the other parameters. The value of the parameter of interest where the profile likelihood is within 1.92 of lmax is the 95% CI.
2.6 Simulated epidemics
Epidemics were simulated in order to test the effect of missing and inaccurate data on parameter estimates. For convenience, we used the Devon demography and the Devon parameter estimates determined by maximum likelihood as the basis for the simulations. Initial conditions were infection of five randomly selected farms. The probability that a farm is infected in a given day is given by equation (2.3). An infected farm is randomly assigned a latent period uniformly distributed from 3 to 7 days. A further 1–3 days, uniformly distributed, then passes before it is reported as an IP. All farms within 1.5 km of an IP are pre-emptively culled (assumed for simplicity). For the first 30 days of the epidemic, both IPs and pre-emptive culls are slaughtered 3 days after reporting. After 30 days, this changes to 1 and 2 days, respectively.
The model is not meant to reflect the control procedures of the UK 2001 epidemic. It is used to compare the changes in estimated parameter values when we degrade epidemic data. For this reason, and because it is difficult to parametrize (Tildesley et al. 2006), we did not model DC culling.
Maximum-likelihood parameter estimates were found using the same likelihood function as for the UK 2001 epidemic. The parameter values used in the simulations are given in table 2. These parameter values give epidemic sizes of a few hundred IPs.
Table 2.
Parameter values used for simulated epidemics.
| parameter | value |
|---|---|
| ρ | 0 or 1 |
| μ | 0 or 1 |
| Tsheep | 11.1×10−6 |
| Tcattle | 6.34×10−6 |
| Scattle | 7.14 |
| δ | 0.71 km |
| b | 1.66 |
2.7 Effect of data degradation on sensitivity and specificity
The effects of missing infections, inaccurate infection dates and constant latent periods on parameter estimates and test sensitivity (probability of accepting the model of changing infectiousness given that it is true) and specificity (probability of accepting the model of constant infectiousness given that it is true) were analysed for two cases: (ρ,μ)=(0,0) and (ρ,μ)=(1,1). For both, 1000 simulations with these parameter values were performed and the model fitted to the resulting epidemics using maximum likelihood.
We tested the effect of missing infections, i.e. unreported infected farms being treated as pre-emptive culls. This could occur in the UK 2001 epidemic when an infected farm was culled before clinical symptoms were apparent because, for example, it was a DC or CP linked to an IP. We consider three cases when unreported infected farms are treated as (i) IPs, (ii) non-infectious IPs, and (iii) pre-emptive culls (as they would be in the UK 2001 epidemic).
We tested the effect of inaccurate infection dates. These were modelled by adjusting the true infection dates of IPs by a number of days randomly chosen from a discrete uniform distribution. We analysed two cases: Uniform(−1,1) and Uniform(−2,2). Unreported infectives were treated as pre-emptive culls.
We tested the effect of assuming equal latent periods for all IPs. Periods of 4–6 days were used (the average latent period in the simulations was 5 days with a range of 3–7 days).
To check the sensitivity of the test for ρ, the model was fitted to each of the 1000 (ρ,μ)=(1,1) simulations both under the full model and with ρ fixed to 0 with all other parameters estimated as usual. The likelihood ratio test was applied to the 1000 pairs of maximum-likelihood estimates to determine significance of ρ. This gave us the frequency that the test correctly accepted the hypothesis that ρ was significantly greater than 0, i.e. sensitivity of the test for ρ=1. A similar procedure was done for μ. Note that for different values of ρ and μ, sensitivity can change. However, the point of the exercise is to demonstrate the qualitative effect of data degradation on sensitivity, not the effect of changes in parameter values.
Test specificity was found in a similar fashion but using the (ρ,μ)=(0,0) simulations and estimating the frequency of non-significant difference from zero.
3. Results
3.1 Parameter estimates from the UK 2001 epidemic
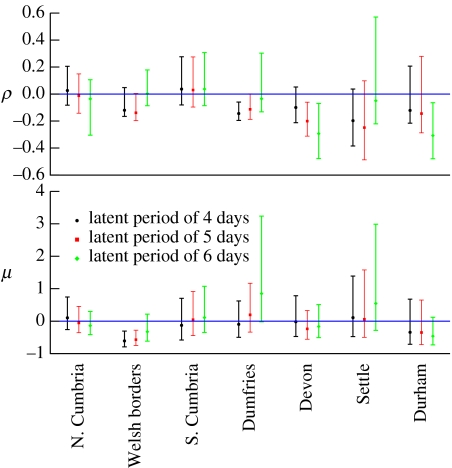
Figure 1 shows the estimated parameter values and 95% CIs of ρ and μ (equation (2.6)) across the various regions and for latent periods of 4 (black circles), 5 (red squares) and 6 days (green diamonds). The other fitted model parameters were consistent with their true values (not shown).
Figure 1.
Maximum-likelihood estimated parameter values and CIs for various regions and for fixed IP latent periods of 4 (black circles), 5 (red squares) and 6 (green diamonds) days. ρ quantifies the rate of change of infectiousness from the day a farm becomes infectious to the day it is reported and μ quantifies infectiousness after reporting. Non-significance from zero suggests no change in infectiousness over infectious period.
3.2 Effects of missing infections, inaccurate infection dates and constant latent periods
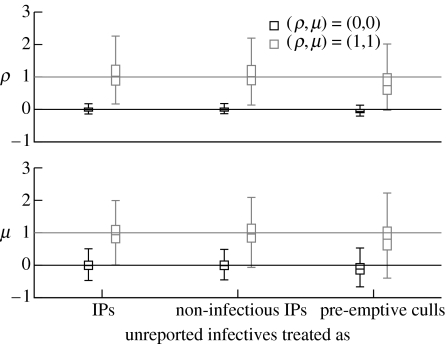
We first tested the effect of missing infections, i.e. unreported infected farms being treated as pre-emptive culls. All farm infection dates and latent periods are known. Figure 2 shows the effect on the parameter estimates when unreported infected farms are treated as (i) IPs, (ii) non-infectious IPs, and (iii) pre-emptive culls. The (ρ,μ)=(0,0) case is shown in black, and the (ρ,μ)=(1,1) case in grey. The boxplots represent the distribution of estimated parameter values found from the simulations.
Figure 2.
Boxplots of the distributions of estimated parameter values from 1000 simulated epidemics. Horizontal line: median; box: 25 and 75 percentiles; whiskers: 1.5 times interquartile range; outliers not shown. Two cases were simulated (ρ,μ)=(0,0) (black) and (ρ,μ)=(1,1) (grey). When estimating the parameter values, infection dates and latent periods of IPs were known. Unreported infectives were treated as IPs (i.e. infectious premises with known infection dates and latent periods), non-infectious IPs (i.e. non-infectious but with known infection dates and latent periods) and pre-emptive culls as they would be for the UK 2001 epidemic (i.e. non-infected and non-infectious).
With full knowledge of the infectiousness of all infected farms (unreported infectives treated as IPs), the median estimated values of ρ and μ are very close to their true values. When unreported infectives are treated as pre-emptive culls, the median estimated values of ρ and μ drop for both cases. This drop is owing to unreported infectives assumed to be uninfected rather than being infected but not infectious, as is shown when we treat unreported infectives as infected but not infectious (non-infectious IPs). The sensitivity of the test when ρ=1 is 85% (table 3), but drops to 57% when unreported infectives are treated as pre-emptive culls. Similarly for μ, the sensitivity drops from 92 to 80%.
Table 3.
Sensitivity and specificity of the significance test for ρ and μ for inaccurate IP infection dates and latent periods.
| type of data degradation | ρ | μ | ||
|---|---|---|---|---|
| sensitivity | specificity | sensitivity | specificity | |
| unreported infectives treated as | ||||
| IPs | 85 | 94 | 92 | 92 |
| non-infectious IPs | 85 | 94 | 92 | 94 |
| pre-emptive culls | 57 | 81 | 80 | 78 |
| td+Uniform(−1,1) | 26 | 81 | 41 | 80 |
| td+Uniform(−2,2) | 19 | 83 | 21 | 79 |
| l=4 days | 89 | 76 | 62 | 59 |
| l=5 days | 77 | 79 | 50 | 65 |
| l=6 days | 55 | 79 | 40 | 79 |
| td+Uniform(−1,1) and l=4 days | 52 | 81 | 38 | 73 |
| td+Uniform(−1,1) and l=5 days | 38 | 82 | 25 | 81 |
| td+Uniform(−1,1) and l=6 days | 26 | 81 | 26 | 85 |
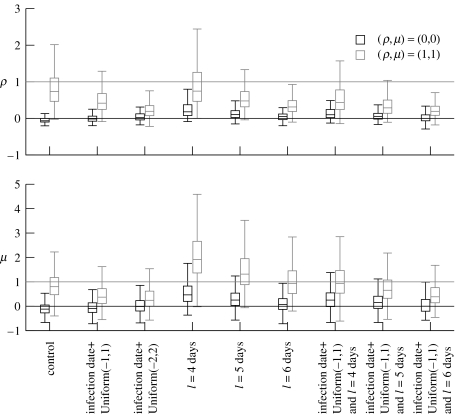
We next tested the effect of inaccurate infection dates. As the error in the infection date increases, the median values of ρ and μ for both cases tend towards zero (figure 3). In other words, the infectiousness tends to be flatter over infectious period when there is uncertainty in infection dates. This is reflected in the test sensitivity which falls to 26% and 19% for ρ and 41% and 21% for μ.
Figure 3.
Boxplots of the distributions of estimated parameter values from 1000 simulated epidemics. Two cases were simulated (ρ,μ)=(0,0) (black) and (ρ,μ)=(1,1) (grey). In all cases, unreported infectives were treated as pre-emptive culls. Control: IP infection dates and latent periods known. IP infection dates (td) were made inaccurate by adjusting them by a number of days uniformly distributed between −1 and 1 or −2 and 2. IP latent periods were assumed fixed at either 4, 5 or 6 days.
Next, we tested the effect of assuming equal latent periods for all IPs. The longer the latent period the lower the estimated values of ρ and μ, and the worse the sensitivity of the test becomes. Specificity of the tests remains around 60–95% for most cases of data degradation.
4. Discussion and conclusion
If we take the parameter estimates in figure 1 as reliable, then in the majority of cases there is no evidence of changing infectiousness over infectious period, i.e. ρ and μ are not significantly different from zero. Some values are significantly lower than zero, suggesting a declining infectiousness. However, even with complete and accurate data, estimates of ρ and μ can be significantly negative even if in reality they are not (for example, the case (ρ,μ)=(0,0) when unreported infectives are treated as IPs in figure 2). This is owing to estimation with finite data and the arbitrariness of statistical significance tests.
However, the conclusions drawn from figure 1 are only valid if we believe the parameter estimates to be reliable. The results from simulations with missing and inaccurate data suggest that the parameter estimates in figure 1 are not reliable. Although we cannot directly compare the estimates from the simulations to the estimates from the UK 2001 epidemic (because the simulations do not model DC and CP culling), we can examine how inaccuracies in the 2001 data qualitatively affect the parameter estimates.
Some farms during the UK 2001 epidemic may have been infected, but were never reported as such because they were pre-emptively culled before clinical signs could appear. It is not yet known how many of these farms there were. In our simulations, such farms do occur and their effect is to reduce estimates of ρ and μ (figure 2), thus reducing the sensitivity of the statistical test (table 3). The infection dates of IPs in the UK 2001 epidemic were usually estimated, either by tracing possible transmission events, or by dating lesions in infected animals. When errors in infection dates are introduced into our simulated data, parameter estimates of ρ and μ tend towards zero (figure 3), again reducing the sensitivity of the test (table 3). This means that infectiousness will show less change over infectious period than in reality. Assuming an equal latent period for all IPs when in reality IP latent period varies also affects the parameter estimates (figure 3), probably more so than missing infections and uncertain infection dates.
In conclusion, our parameter estimates of the trends in infectiousness during the UK 2001 epidemic are not robust to uncertainties in IP infection and infectious status. Having said that, the majority of our estimates are not significantly different from zero. This means that allowing infectiousness to change over infectious period does not significantly improve the fit of our model to the available data. Parsimony then dictates that we must assume a constant infectiousness until evidence to the contrary arises.
All models developed during the epidemic agreed that the control policy as implemented was not controlling the epidemic, and that a fully implemented IP/CP cull would if there were no increases in infectiousness over infectious period. If infectiousness did increase over infectious period, however, then CP culling may or may not have been effective depending on the magnitude by which infectiousness increased. Such uncertainty in this and other parameter estimates means that it is impossible to find an optimal control strategy (in terms of numbers of animals killed) that includes neighbourhood culling (Matthews et al. 2003). However, it has been shown that total losses are not sensitive for a control effort above the optimal, but can quickly increase below the optimal (Matthews et al. 2003). Our results suggest, therefore, that CP culling was a less risky policy than IP and DC culling alone in the face of considerable uncertainty in the transmission process.
The likelihood function we used to fit the model to the data could be expanded in several ways, and may affect our parameter estimates but not our conclusions about data degradation. We are currently working on estimating infection dates, latent periods and the proportion of IPs and DCs infected using Markov chain Monte Carlo-based Bayesian inference.
Acknowledgments
This research is supported by the Wellcome Trust. We thank Miles Thomas from the Central Science Laboratory, DEFRA, Sand Hutton, Yorkshire for his invaluable help with the data.
References
- Anderson I. The Stationary Office; London, UK: 2002. Foot and mouth disease 2001: lessons to be learned inquiry. [Google Scholar]
- Ap Dewi A, Molina-Flores B, Edwards-Jones G. A generic spreadsheet model of a disease epidemic with application to the first 100 days of the 2001 outbreak of foot-and-mouth disease in the UK. Vet. J. 2004;167:167–174. doi: 10.1016/S1090-0233(03)00149-7. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Donnelly C.A, Anderson R.M. The foot-and-mouth epidemic in Great Britain: pattern of spread and impact of interventions. Science. 2001a;292:1155–1160. doi: 10.1126/science.1061020. [DOI] [PubMed] [Google Scholar]
- Ferguson N.M, Donnelly C.A, Anderson R.M. Transmission intensity and impact of control policies on the foot and mouth epidemic in Great Britain. Nature. 2001b;414:542–548. doi: 10.1038/35097116. [DOI] [PubMed] [Google Scholar]
- Ferris N.P, King D.P, Reid S.M, Shaw A.E, Hutchings G.H. Comparisons of original laboratory results and retrospective analysis by real-time reverse transcriptase-PCR of virological samples collected from confirmed cases of foot-and-mouth disease in the UK in 2001. Vet. Rec. 2006;159:373–378. doi: 10.1136/vr.159.12.373. [DOI] [PubMed] [Google Scholar]
- Gibbens J.C, Sharpe C.E, Wilesmith J.W, Mansley L.M, Michalopoulou E, Ryan J.B.M, Hudson M. Descriptive epidemiology of the 2001 foot-and-mouth disease epidemic in Great Britain: the first five months. Vet. Rec. 2001;149:729–743. [PubMed] [Google Scholar]
- Hyslop N.S.G. Secretion of foot-and-mouth disease virus and antibody in the saliva of infected and immunized cattle. J. Comp. Path. 1965;75:111–117. doi: 10.1016/0021-9975(65)90001-0. [DOI] [PubMed] [Google Scholar]
- Kao R.R. Landscape fragmentation and foot-and-mouth disease. Vet. Rec. 2001;148:746–747. doi: 10.1136/vr.148.24.746. [DOI] [PubMed] [Google Scholar]
- Kao R.R. The impact of local heterogeneity on alternative control strategies for foot-and-mouth disease. Proc. R. Soc. B. 2003;270:2557–2564. doi: 10.1098/rspb.2003.2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M.J, et al. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science. 2001;294:813–817. doi: 10.1126/science.1065973. [DOI] [PubMed] [Google Scholar]
- Matthews L, Haydon D.T, Shaw D.J, Chase-Topping M.E, Keeling M.J, Woolhouse M.E.J. Neighbourhood control policies and the spread of infectious diseases. Proc. R. Soc. B. 2003;270:1659–1666. doi: 10.1098/rspb.2003.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press W.H, Flannery B.P, Teukolsky S.A, Vetterling W.T. 2nd edn. Cambridge University Press; Cambridge, UK: 1992. Numerical recipes in C: the art of scientific computing. [Google Scholar]
- Tildesley M.J, Savill N.J, Shaw D.J, Deardon R, Brooks S.P, Woolhouse M.E.J, Grenfell B.T, Keeling M.J. Optimal reactive vaccination strategies for a foot-and-mouth outbreak in the UK. Nature. 2006;440:83–86. doi: 10.1038/nature04324. [DOI] [PubMed] [Google Scholar]