Abstract
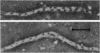
A new temperature-sensitive mutant strain of Escherichia coli K-12 which forms filaments at 42 C has been described. The mutant, Y16, maintained growth and deoxyribonucleic acid synthesis at 42 C. The resulting multinucleate filaments gradually lost their viability at 42 C but could be recovered, even after 240 min of incubation, upon return to 30 C. Septation was resumed and growth was promptly re-established at normal rates. Recovery still took place in the presence of chloramphenicol added to the culture at the time of temperature shift from 42 to 30 C. A study has been made of the effects of adenine and various nucleosides on cultures of strain Y16 as compared with another filament-forming mutant, T44 tif-. Adenine (75 mug/ml), known to promote filamentation of strain T44 tif-, prevented the development of filaments and the loss of viability in cultures of Y16. Recovery of septation after temperature shift in cultures containing adenine presented a pattern similar to that found with the adenine-less cultures. Protection afforded by adenine at 42 C could be reversed by the addition of guanosine plus cytidine (100 mug/ml each). The effects of high concentrations of adenine and nucleosides on strain Y16 thus are the reverse of those observed with mutant T44 tif-. However, whereas tif-1 mutation promotes prophage induction at restrictive temperatures, no modification could be detected in the process of prophage induction in cultures of the lambda-lysogenic derivative of Y16 at 42 C, be it spontaneous or ultraviolet-mediated induction. The osmolarity increase afforded by 1% NaCl added to the medium did not alter the phenotype characteristics of strain Y16. The mutation has been mapped between argG and bgl. A close linkage has been observed between ftsH and argG, thereby locating the new mutation near 61 min on the map of E. coli chromosome, a previously undescribed region involved in cell division. The evidence reported indicates that strain Y16 differs in several respects from the already descirbed strains of the same class.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. S., Filip C. C., Gustafson R. A., Allen R. G., Walker J. R. Regulation of bacterial cell division: genetic and phenotypic analysis of temperature-sensitive, multinucleate, filament-forming mutants of Escherichia. J Bacteriol. 1974 Mar;117(3):978–986. doi: 10.1128/jb.117.3.978-986.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen R. G., Smith J. A., Knudsen R. C., Walker J. R. Initial characterization of temperature-sensitive cell division mutants of Escherichia coli. Biochem Biophys Res Commun. 1972 Jun 9;47(5):1074–1079. doi: 10.1016/0006-291x(72)90943-6. [DOI] [PubMed] [Google Scholar]
- Castellazzi M., George J., Buttin G. Prophage induction and cell division in E. coli. I. Further characterization of the thermosensitive mutation tif-1 whose expression mimics the effect of UV irradiation. Mol Gen Genet. 1972;119(2):139–152. doi: 10.1007/BF00269133. [DOI] [PubMed] [Google Scholar]
- GOLDTHWAIT D., JACOB F. SUR LE M'ECANISME DE L'INDUCTION DU D'EVELOPPEMENT DU PROPHAGE CHEZ LES BACT'ERIES LYSOG'ENES. C R Hebd Seances Acad Sci. 1964 Jul 20;259:661–664. [PubMed] [Google Scholar]
- Hirota Y., Ryter A., Jacob F. Thermosensitive mutants of E. coli affected in the processes of DNA synthesis and cellular division. Cold Spring Harb Symp Quant Biol. 1968;33:677–693. doi: 10.1101/sqb.1968.033.01.077. [DOI] [PubMed] [Google Scholar]
- Kirby E. P., Jacob F., Goldthwait D. A. Prophage induction and filament formation in a mutant strain of Escherichia coli. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1903–1910. doi: 10.1073/pnas.58.5.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohiyama M., Cousin D., Ryter A., Jacob F. Mutants thermosensibles d'Escherichia coli K 12. I. Isolement et caractérisation rapide. Ann Inst Pasteur (Paris) 1966 Apr;110(4):465–486. [PubMed] [Google Scholar]
- LATARJET R., MORENNE P., BERGER R. Un appareil simple pour le dosage des rayonnements ultraviolets émis par les lampes germicides. Ann Inst Pasteur (Paris) 1953 Aug;85(2):175–184. [PubMed] [Google Scholar]
- LOW B., WOOD T. H. A QUICK AND EFFICIENT METHOD FOR INTERRUPTION OF BACTERIAL CONJUGATION. Genet Res. 1965 Jul;6:300–303. doi: 10.1017/s001667230000416x. [DOI] [PubMed] [Google Scholar]
- LURIA S. E., BURROUS J. W. Hybridization between Escherichia coli and Shigella. J Bacteriol. 1957 Oct;74(4):461–476. doi: 10.1128/jb.74.4.461-476.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MASON D. J., POWELSON D. M. Nuclear division as observed in live bacteria by a new technique. J Bacteriol. 1956 Apr;71(4):474–479. doi: 10.1128/jb.71.4.474-479.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Kaneko H., Tamura G. Thermosensitive mutant of Escherichia coli requiring new protein synthesis to recover cellular division ability. Biochem Biophys Res Commun. 1971 Feb 19;42(4):669–675. doi: 10.1016/0006-291x(71)90540-7. [DOI] [PubMed] [Google Scholar]
- Nagai K., Tamura G. Mutant of Escherichia coli with thermosensitive protein in the process of cellular division. J Bacteriol. 1972 Nov;112(2):959–966. doi: 10.1128/jb.112.2.959-966.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Groves D. J., Clark D. J. Regulation of Cell Division in Escherichia coli: Characterization of Temperature-Sensitive Division Mutants. J Bacteriol. 1970 Dec;104(3):1052–1064. doi: 10.1128/jb.104.3.1052-1064.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard M., Hirota Y. Effet des sels sur le processus de division cellulaire d'E. coli. C R Acad Sci Hebd Seances Acad Sci D. 1969 Mar 3;268(9):1335–1338. [PubMed] [Google Scholar]
- Ricard M., Hirota Y. Process of cellular division in Escherichia coli: physiological study on thermosensitive mutants defective in cell division. J Bacteriol. 1973 Oct;116(1):314–322. doi: 10.1128/jb.116.1.314-322.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff W., Kirby E. P., Goldthwait D. A. Cell division and prophage induction in Escherichia coli: studies of nucleotide levels. J Bacteriol. 1971 Jun;106(3):994–1004. doi: 10.1128/jb.106.3.994-1004.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M., Schaechter M. Control of cell division in bacteria. Bacteriol Rev. 1974 Jun;38(2):199–221. doi: 10.1128/br.38.2.199-221.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone A. B. Regulation of cell division in a temperature-sensitive division mutant of Escherichia coli. J Bacteriol. 1973 Nov;116(2):741–750. doi: 10.1128/jb.116.2.741-750.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. L., Trotter C. D. Linkage map of Escherichia coli strain K-12. Bacteriol Rev. 1972 Dec;36(4):504–524. doi: 10.1128/br.36.4.504-524.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAN DE PUTTE P., VAN DILLEWIJN, ROERSCH A. THE SELECTION OF MUTANTS OF ESCHERICHIA COLI WITH IMPAIRED CELL DIVISION AT ELEVATED TEMPERATURE. Mutat Res. 1964 Jul;106:121–128. doi: 10.1016/0027-5107(64)90014-4. [DOI] [PubMed] [Google Scholar]
- Zusman D. R., Inouye M., Pardee A. B. Cell division in Escherichia coli: evidence for regulation of septation by effector molecules. J Mol Biol. 1972 Aug 14;69(1):119–136. doi: 10.1016/0022-2836(72)90027-7. [DOI] [PubMed] [Google Scholar]