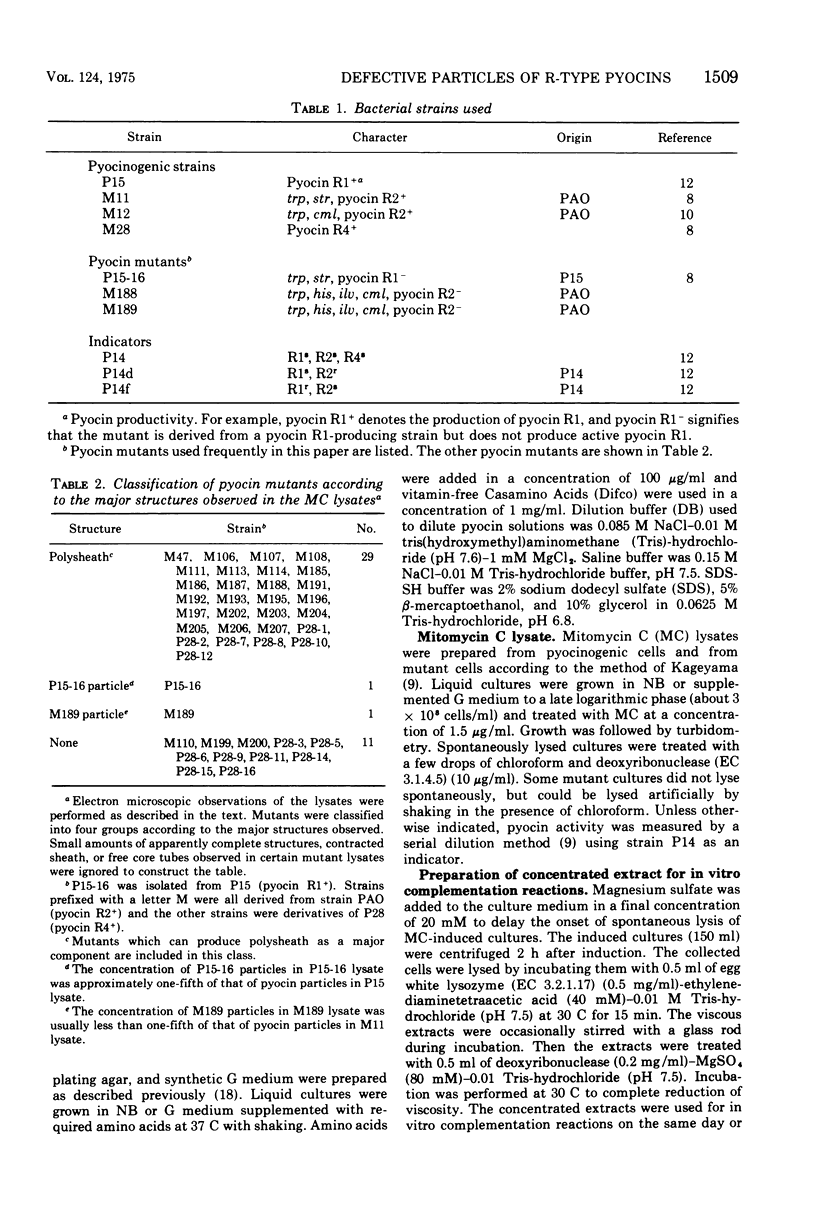
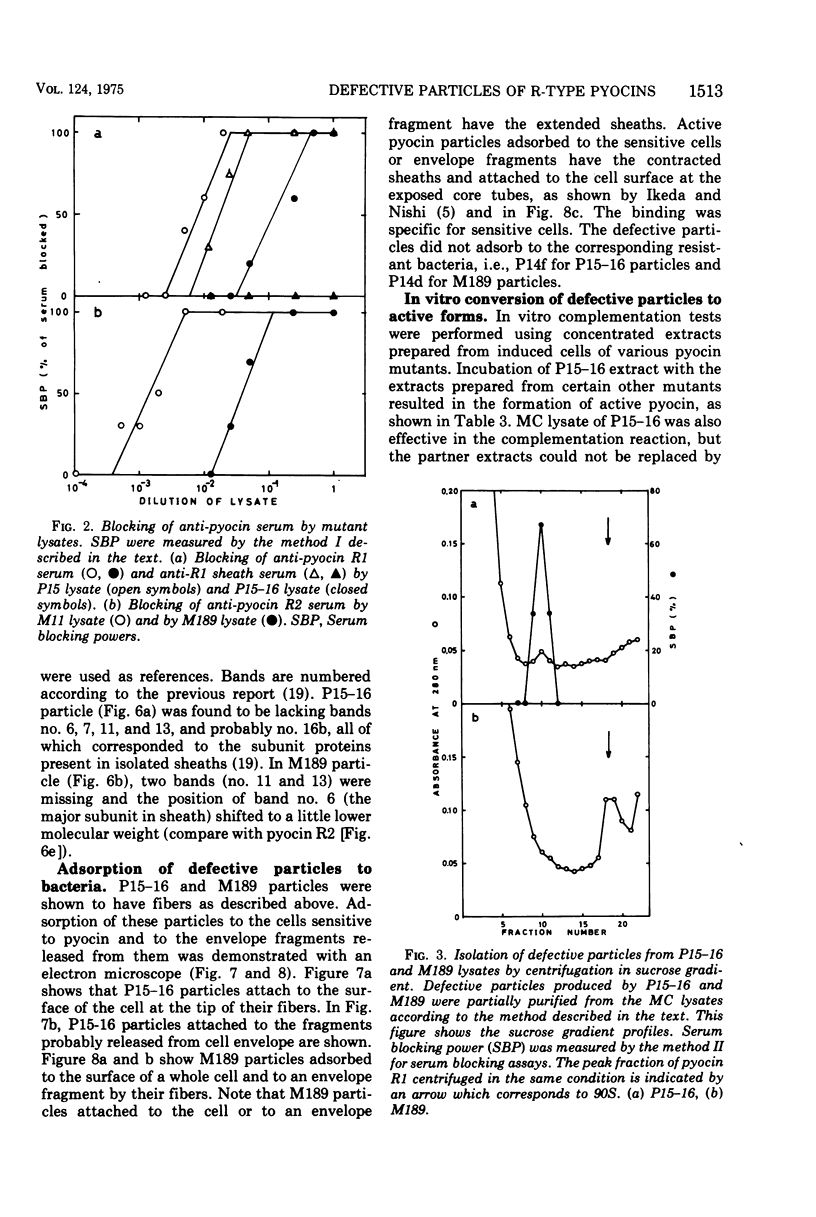
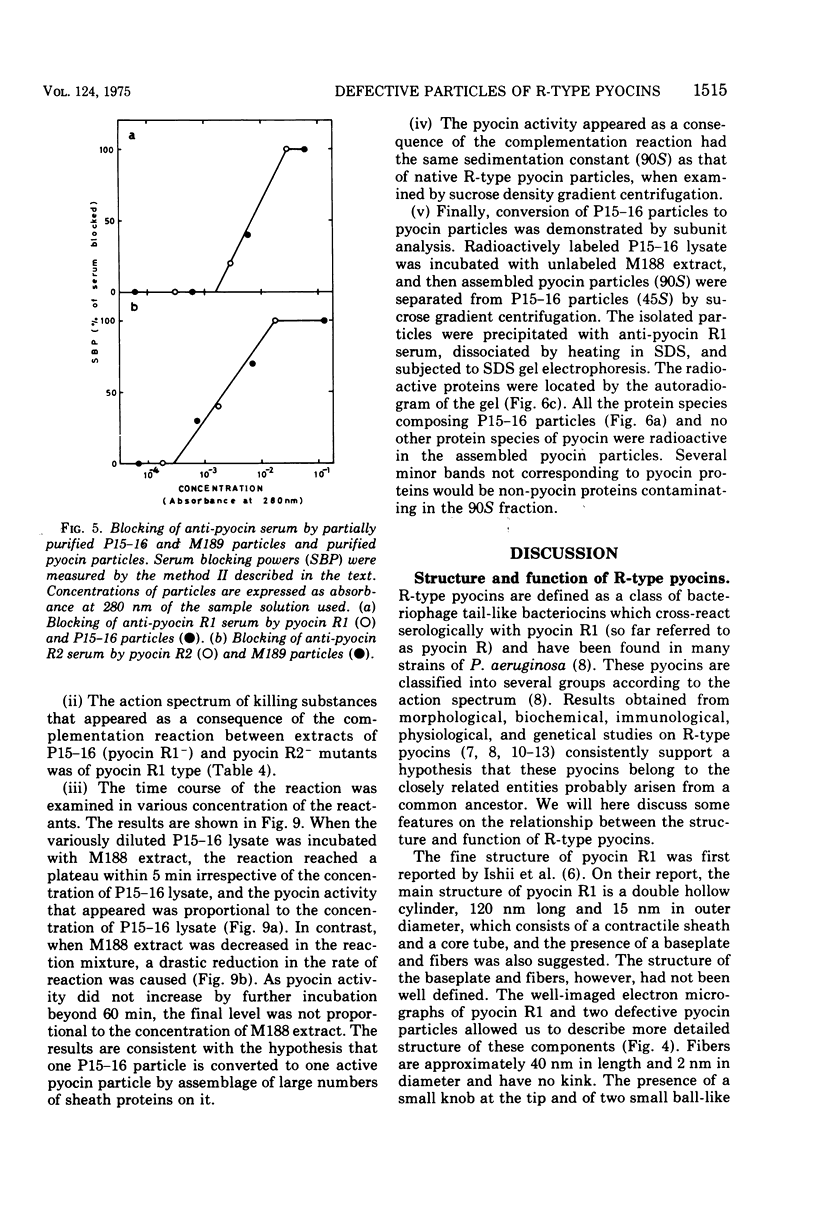
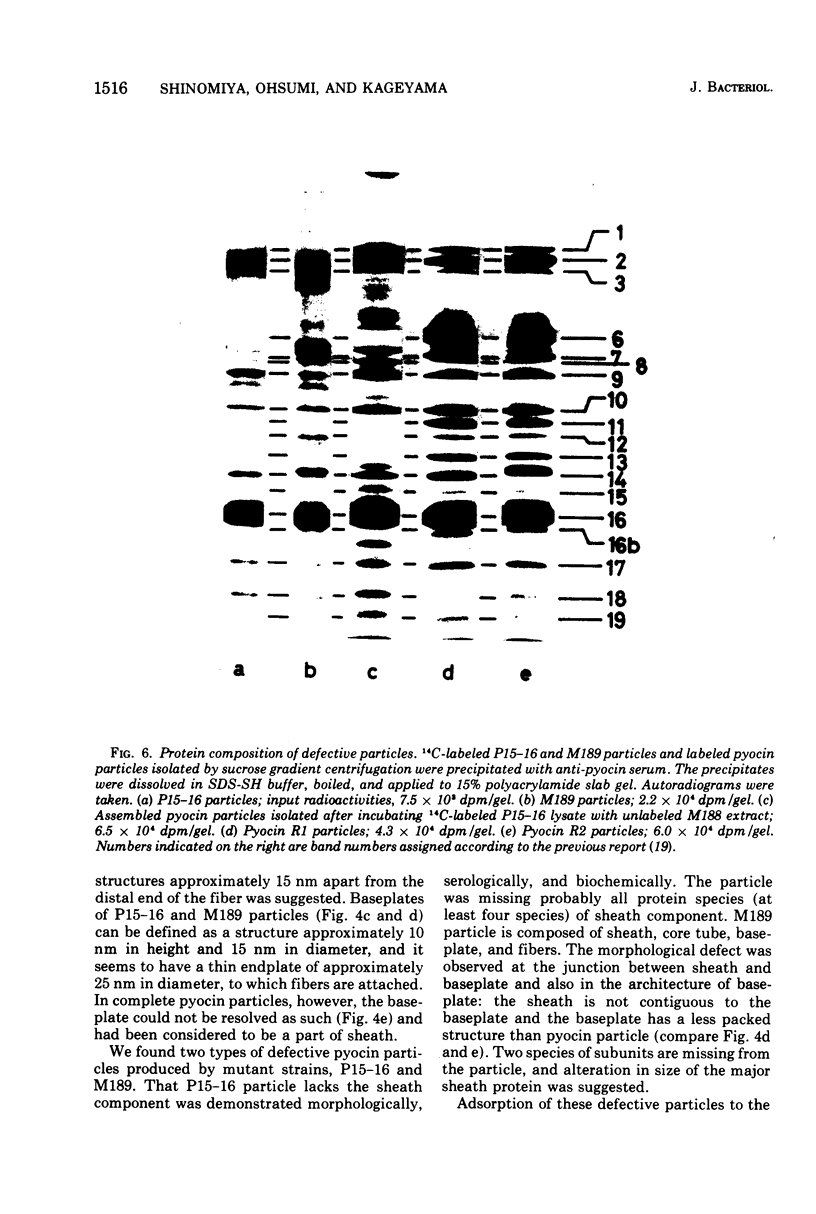
Abstract
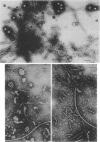
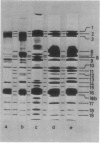
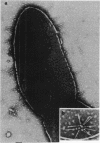
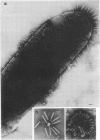
Mutants of Pseudomonas aeruginosa, defective in the production of active R-type pyocins, were isolated from pyocinogenic strains and their products were characterized. Polysheath-like structures were found in induced lysates of 29 out of 42 mutants. Two mutants (strain P15-16 and M189) were found to produce special defective particles, which were characterized in detail. The other 11 mutants did not produce significant amounts of any structure visible under an electron microscope. Serum blocking powers were found in lysates from P15-16 and M189 to significant amounts. Defective particle produced by strain P15-16 lacked the sheath component, whereas M189 had morphological defects at the junction between sheath and baseplate, and also in the architecture of baseplate. Both defective particles could adsorb to the surface of bacteria, that were sensitive to pyocin, at the tip of their fibers without killing cells. All M189 particles attached to the bacteria had the extended sheaths. Therefore, attachment to the bacteria by fibers is not sufficient to kill cells, and contraction of sheath must occur after the initial adsorption by fibers for pyocin to express its biological activity. Defective particles of strain P15-16, which was derived from strain P15 (a pyocin R1 producer), could be converted to active forms by an in vitro complementation reaction with extracts from certain mutants originated from strain PAO (a pyocin R2 producer). This result indicated the exchangeability of components between R-type pyocins belonging to the different groups.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BRENNER S., HORNE R. W. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta. 1959 Jul;34:103–110. doi: 10.1016/0006-3002(59)90237-9. [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Lielausis I. Serological studies with mutants of phage T4D defective in genes determining tail fiber structure. Genetics. 1965 Dec;52(6):1187–1200. doi: 10.1093/genetics/52.6.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S., Wood W. B. Morphogenesis of bacteriophage T4 in extracts of mutant-infected cells. Proc Natl Acad Sci U S A. 1966 Mar;55(3):498–505. doi: 10.1073/pnas.55.3.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HORI T., ITASAKA O., HASHIMOTO T., INOUE H. BIOCHEMISTRY OF SHELL-FISH LIPID. II. ISOLATION OF THE ETHANOLAMINE-CONTAINING SPHINGOLIPID FROM CORBICULA, CORBICULA SANDAI. J Biochem. 1964 May;55:545–552. [PubMed] [Google Scholar]
- Higerd T. B., Baechler C. A., Berk R. S. In vitro and in vivo characterization of pyocin. J Bacteriol. 1967 Jun;93(6):1976–1986. doi: 10.1128/jb.93.6.1976-1986.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S. I., Nishi Y., Egami F. The fine structure of a pyocin. J Mol Biol. 1965 Sep;13(2):428–431. doi: 10.1016/s0022-2836(65)80107-3. [DOI] [PubMed] [Google Scholar]
- KELLENBERGER E., ARBER W. Electron microscopical studies of phage multiplication. I. A method for quantitative analysis of particle suspensions. Virology. 1957 Apr;3(2):245–255. doi: 10.1016/0042-6822(57)90091-0. [DOI] [PubMed] [Google Scholar]
- Kaziro Y., Tanaka M. Studies on the mode of action of pyocin. I. Inhibition of macromolecular synthesis in sensitive cells. J Biochem. 1965 May;57(5):689–695. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Shinomiya T., Egami F. Preferential inhibition of pyocin R production by dihydrostreptomycin. J Biochem. 1967 Dec;62(6):679–687. doi: 10.1093/oxfordjournals.jbchem.a128724. [DOI] [PubMed] [Google Scholar]
- Shinomiya T. Studies on biosynthesis and morphogenesis of R-type pyocins of Pseudomonas aeruginosa. 3. Subunits of pyocin R and their precipitability by anti-pyocin R serum. J Biochem. 1972 Sep;72(3):499–510. doi: 10.1093/oxfordjournals.jbchem.a129929. [DOI] [PubMed] [Google Scholar]
- Shinomiya T. Studies on biosynthesis and morphogenesis of R-type pyocins of Pseudomonas aeruginosa. II. Biosynthesis of antigenic proteins and their assembly into pyocin particles in mitomycin C-induced cells. J Biochem. 1972 Jul;72(1):39–48. doi: 10.1093/oxfordjournals.jbchem.a129895. [DOI] [PubMed] [Google Scholar]
- Shinomiya T. Studies on the biosynthesis and morphogenesis of R-type pyocins of Pseudomonas aeruginosa. IV. Quantitative estimation of the rates of synthesis of pyocin R subunit proteins. J Biochem. 1974 Nov;76(5):1083–1094. [PubMed] [Google Scholar]
- Simon L. D., Anderson T. F. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology. 1967 Jun;32(2):279–297. doi: 10.1016/0042-6822(67)90277-2. [DOI] [PubMed] [Google Scholar]
- Simon L. D., Swan J. G., Flatgaard J. E. Functional defects in T4 bacteriophages lacking the gene 11 and gene 12 products. Virology. 1970 May;41(1):77–90. doi: 10.1016/0042-6822(70)90056-5. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Luftig R. B., Wood W. B. Interaction of bacteriophage T4 tail fiber components with a lipopolysaccharide fraction from Escherichia coli. J Mol Biol. 1970 Jul 28;51(2):423–434. doi: 10.1016/0022-2836(70)90152-x. [DOI] [PubMed] [Google Scholar]
- Yui C. Structure of pyocin R. I. Isolation of sheath from pyocin R by alkali treatment and its properties. J Biochem. 1971 Jan;69(1):101–110. doi: 10.1093/oxfordjournals.jbchem.a129437. [DOI] [PubMed] [Google Scholar]