Abstract
Previously, renal cell carcinoma tissues were reported to display a marked reduction of components of the respiratory chain. To elucidate a possible relationship between tumourigenesis and alterations of oxidative phosphorylation, we screened for mutations of the mitochondrial DNA (mtDNA) in renal carcinoma tissues and patient-matched normal kidney cortex. Seven of the 15 samples investigated revealed at least one somatic heteroplasmic mutation as determined by denaturating HPLC analysis (DHPLC). No homoplasmic somatic mutations were observed. Actually, half of the mutations presented a level of heteroplasmy below 25%, which could be easily overlooked by automated sequence analysis. The somatic mutations included four known D-loop mutations, four so far unreported mutations in ribosomal genes, one synonymous change in the ND4 gene and four nonsynonymous base changes in the ND2, COI, ND5 and ND4L genes. One renal cell carcinoma tissue showed a somatic A3243G mutation, which is a known frequent cause of MELAS syndrome (mitochondrial encephalomyopathy, lactic acidosis, stroke-like episode) and specific compensatory alterations of enzyme activities of the respiratory chain in the tumour tissue. No difference between histopathology and clinical progression compared to the other tumour tissues was observed. In conclusion, the low abundance as well as the frequently observed low level of heteroplasmy of somatic mtDNA mutations indicates that the decreased aerobic energy capacity in tumour tissue seems to be mediated by a general nuclear regulated mechanism.
Keywords: oxidative phosphorylation, mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes, denaturing HPLC, mitochondrial DNA mutations, renal cell carcinoma
Renal cell carcinoma is the most common malignancy arising in the adult kidney. Already five decades ago it was shown that the vast majority of tumours display a high rate of glycolysis under aerobic conditions (Warburg, 1956). Accordingly, in renal cell carcinomas, an increase of proteins involved in major steps of the glycolytic pathway and a decrease of the gluconeogenic reactions was observed (Faure Vigny et al, 1996; Unwin et al, 2003). Furthermore, a depletion of the activity of several mitochondrial enzymes was demonstrated (Faure Vigny et al, 1996; Unwin et al, 2003; Meierhofer et al, 2004). Human solid tumours endure profound hypoxia, which indicates that adaptation to hypoxic conditions is a crucial step in tumour progression. Recently, it was postulated that deficiency of the von Hippel Lindau (VHL) protein, which is observed in most renal carcinomas, could be one of the factors responsible for downregulation of the biogenesis of complexes of the oxidative phosphorylation (OXPHOS) (Shiao et al, 2000; Hervouet et al, 2005).
Since some subunits of the OXPHOS are encoded by mitochondrial DNA (mtDNA), alterations of mtDNA may influence OXPHOS activity. Mitochondrial DNA has a high mutation rate due to the damage produced by free radicals, the lack of protective action by histones and the limited capacity of repair of the mtDNA (Brown et al, 1979; Pettepher et al, 1991). The mutation rate of mtDNA has been reported to be as much as two orders of magnitude greater than that of nuclear DNA (Khrapko et al, 1997). A mutation in mtDNA expands either partially (heteroplasmy) or totally replaces all mtDNA (homoplasmy). However, it is still unclear how mutated mtDNA expands in cells.
Recently, a high incidence of specific mtDNA alterations has been reported for gastric (Maximo et al, 2001; Wu et al, 2005), prostate (Jeronimo et al, 2001; Petros et al, 2005), pancreatic (Jones et al, 2001), skin (Girald-Rosa et al, 2005), colorectal (Polyak et al, 1998; Hibi et al, 2001a; Lievre et al, 2005), urinary bladder (Fliss et al, 2000), thyroid (Yeh et al, 2000), oesophageal (Hibi et al, 2001b; Kumimoto et al, 2004), liver (Nishikawa et al, 2001), breast (Richard et al, 2000; Tan et al, 2002; Zhu et al, 2005), uterine cancers (Pejovic et al, 2004) as well as chromophobe renal cell carcinoma (Nagy et al, 2002). Of all mtDNA mutations reported in cancer tissues, only a few are known to be of pathological relevance as shown for patients with disorders of the mitochondrial energy metabolism. For example, a G5521A mutation, which is known to cause a late onset mitochondrial myopathy (Silvestri et al, 1998), was detected in a lung cancer tissue (Fliss et al, 2000). A mutation (G13708A) reported in patients with Leber's hereditary optic neuropathy (LOHN) (Brown et al, 1992) was found in a breast cancer tissue (Parrella et al, 2001) and a mutation typical for mitochondrial encephalomyopathy with lactic acidosis and stroke-like episodes (MELAS) was detected in a colon cancer sample (Lorenc et al, 2003). As would be anticipated, these patients, who all were carrying only tumour-specific pathogenic mtDNA mutations, did not show clinical signs of mtDNA-related disease. Recently, it was shown that the introduction of a pathogenic mtDNA mutations, for example, in the mitochondrial-encoded ATPase 6 gene (MTATP6), can lead to declined respiration and accelerated growth of tumour cells via inhibition of apoptosis (Petros et al, 2005; Shidara et al, 2005).
To determine whether the downregulation of the enzyme activities reported in renal carcinoma is associated with mtDNA mutations, we screened the entire mtDNA of 15 paired tumour and the corresponding normal kidney samples by denaturating HPLC analysis (DHPLC) analysis. Occurrence and type of somatic mtDNA mutations were compared to the enzymatic activity of respiratory chain complexes.
MATERIALS AND METHODS
Patients
Tumour and the corresponding healthy cortex tissue from 15 patients were obtained by nephrectomy at the Department of Urology, Salzburg as previously reported (Meierhofer et al, 2004). The tumour classification was performed according to Storkel et al (1997). DNA was isolated by proteinase K digestion followed by phenol/chloroform extraction.
PCR amplification of entire mitochondrial genome and mutation analysis
The whole human mtDNA of all 15 patients was amplified using 48 overlapping PCR fragments. Mutation detection by DHPLC was performed as reported previously (Meierhofer et al, 2005). As estimated from the areas under the peaks of the DHPLC chromatogram, the degree of heteroplasmy was divided into lower 25% (<25%), between 25 and 75% (25–75%) and over 75% (>75%). All samples showing a homo- and heteroduplex peak of the same extent in DHPLC analysis were sequenced directly. Heteroplasmic peaks lower than the homoplasmic peak were manually collected and reamplified as previously reported (Meierhofer et al, 2005).
Mutations were analysed with the Beckman software investigator (Fullerton, CA, USA). Exact positions of mutations and amino-acid changes were defined with the mitoAnalyser tool (MitoAnalyzer, 2000), using the mtDNA genbank sequence J01415.1 as reference.
Determination of mutational load of the A3243G mutation
A PCR fragment was amplified with the following primers: forward, 5′-TCCCTGTACGAAAGGACAAGA-3′; reverse, 5′-AGGAGTAGGAGGTTGGCCAT-3′ and with cycling and PCR conditions as reported previously (Meierhofer et al, 2005). The last PCR cycle was followed by a denaturating and reannealing step. Restriction digestion with HaeIII, which digests only the mutation-specific restriction side 3243G and one control restriction side on each side of the PCR fragment, was performed. The percentage of mutational load was detected by densitometry of ethidium-bromide-stained agarose gels (Uziel et al, 1997; van den Bosch et al, 2004).
Enzyme measurements
The following enzyme activities: citrate synthase, complex II, complex IV and oligomycin-sensitive ATPase activity of complex V were determined and reported previously (Meierhofer et al, 2004). In addition, complex I activity was measured according to Rustin et al (1994).
Western blot analysis of the VHL tumour suppressor protein
After separation of the 600 g homogenate (Meierhofer et al, 2004), 18 μg protein per lane were loaded on a 10% polyacrylamide gel and Western blot analysis was performed according to Berger et al (2003). The following antibodies were used: mouse monoclonal antibody against VHL protein (Cat# 556347; BD Bioscience, Palo Alto, CA, USA; 1 μg/ml), alkaline phosphatase-conjugated rabbit anti-mouse immunoglobulins (Dako, Golstrup, Denmark; 1 : 5000).
RESULTS
MtDNA mutations
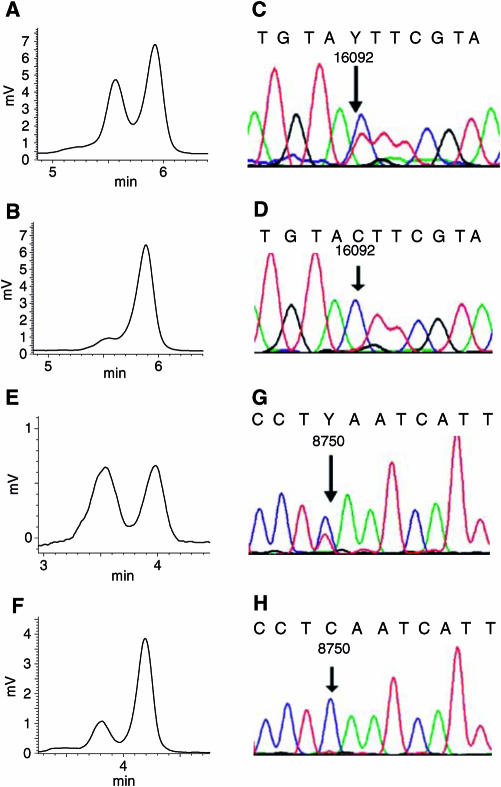
The mutation screening of the entire mtDNA of 15 primary renal carcinoma and matched control kidney cortex tissues was carried out by DHPLC analysis. Five matched tumour/kidney pairs showed one to two heteroplasmic mutations (six D-loop, one 16s rRNA and one COI) of the mtDNA with no difference of the degree of heteroplasmy in the two tissues (Table 1). Interestingly, in two patients, a shift in heteroplasmy from the kidney to the corresponding tumour tissue was observed (cases 11, 12: Table 1; Figure 1). Loss of low-level heteroplasmy present in normal cortex tissue was observed in cases 4 (310 C insertion; A8483G) and 14 (T72C). Only three conventional renal carcinomas with grades 1, 2 and 4 carried neither a somatic nor a heteroplasmic mtDNA mutation.
Table 1. Nonsomatic heteroplasmies in matched tissue pairs.
| Case | Tumour type | Grade | Nucleotide position | Heteroplasmya in kidney/tumour (%) | Gene | Amino acid | Notes |
|---|---|---|---|---|---|---|---|
| 6 | Papillary RC | 2 | A16206G | >75 | D-loop | d | |
| 6 | C16348T | >75 | D-loop | d | |||
| 7 | Sarkomatoid RC | 4 | A16206G | 25–75 | D-loop | d | |
| 7 | C16348T | 25–75 | D-loop | d | |||
| 11 | Conv. RCb | 3 | T16092C | from 40 to >95 | D-loop | c | |
| 12 | Conv. RC | 2 | T8750C | from 50 to 80 | ATP6 | L75S | d |
| 13 | Conv. RC | 1/2 | T1809C | >75 | 16S rRNA | d | |
| 13 | G6260A | 25–75 | COI | syn | c | ||
| 14 | Papillary RC | 1 | G75A | 25–75 | D-loop | c | |
| 15 | Conv. RC | 1 | T16093C | 25–75 | D-loop | c |
Estimated by denaturating HPLC analysis.
Conv.=conventional; RC=renal carcinoma; syn=synonymous.
c=Previously reported polymorphism (MITOMAP database).
d=Novel mutation.
Figure 1.
Analysis of a shift of heteroplasmy in matched tissue pairs. Denaturating HPLC analysis of a PCR fragment (15587–16185) from case 11 at 56.0°C oven temperature (A, B). Denaturating HPLC analysis revealed a heteroplasmy of the kidney tissue of about 40% (A) and of the corresponding renal carcinoma tissue of over 95% (B). Sequence analysis indicates a 16092C/T heteroplasmy in the kidney tissue (C) and the 16092C mutation in the corresponding renal carcinoma (D). Denaturating HPLC analysis of a PCR fragment (8466–8925) from case 12 at 57.8°C oven temperature (E, F). The kidney tissue shows a heteroplasmy of about 50% (E) and the corresponding carcinoma tissue of about 80% (F). Sequence analysis revealed a 8750C/T heteroplasmy in the kidney tissue (G) and the 8750C variation in the corresponding carcinoma tissue (H). Y=C+T.
In total, 14 types of somatic genetic mutations were detected in seven patients (Table 2). No somatic homoplasmic mutations were detected in any of these 15 renal carcinoma tissues. Notably, the level of heteroplasmy of somatic mutations was frequently below 25% (Table 2), whereas all nonsomatic heteroplasmies were above 25% (Table 1).
Table 2. Gain of somatic mutations of the entire mitochodrial DNA genome in renal carcinoma tissues.
| Case | Tumour type | Grade | Nucleotide position | Heteroplasmya in tumour (%) | Gene | Amino acid | Notes |
|---|---|---|---|---|---|---|---|
| 1 | Conv. RCb | 2/3 | C338T | <25 | D-loop | c | |
| 1 | A1578G | <25 | 12S rRNA | d | |||
| 1 | G12007A | <25 | ND4 | syn | c | ||
| 2 | Conv. RC | 3 | G4584A | <25 | ND2 | A39T | d |
| 3 | Conv. RC | 2 | G94A | >75 | D-loop | c | |
| 3 | A7423G | <25 | COI | E507G | d | ||
| 4 | Conv. RC | 3 | T204C | 25–75 | D-loop | c | |
| 4 | A3243G | >75 | tRNA LEU(UUR) | MELAS | |||
| 4 | G1169A | >75 | 12S rRNA | d | |||
| 4 | C12510A | >75 | ND5 | D58E | d | ||
| 5 | Papillary RC | 1 | T2222C | 25–75 | 16S rRNA | d | |
| 6 | Papillary RC | 2 | C1566T | <25 | 12S rRNA | d | |
| 6 | T10579C | <25 | ND4L | M37T | d | ||
| 7 | Sarkomatoid RC | 4 | C16174T | 25–75 | D-loop | c |
Estimated by denaturating HPLC analysis.
Conv.=conventional; RC=renal carcinoma; syn=synonymous.
cPreviously reported polymorphism (MITOMAP database).
dNovel mutation.
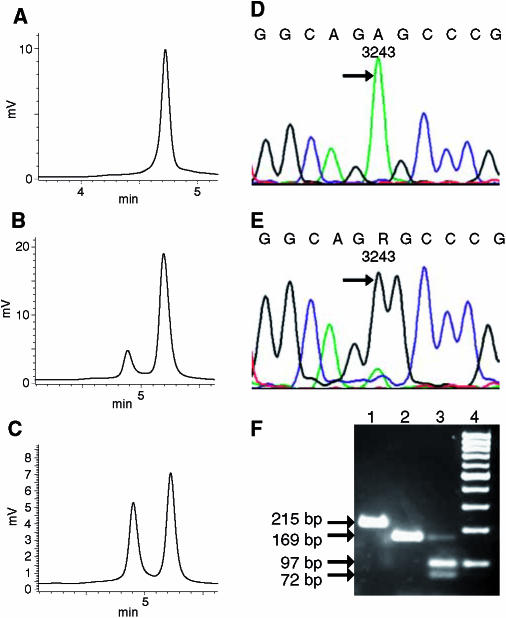
The somatic mutations included four known D-loop mutations, four so far unreported mutations in ribosomal genes, one synonymous change in the ND4 gene and four nonsynonymous base changes in the ND2, COI, ND5 and ND4L genes (Table 2). Interestingly, in one tumour tissue, the tRNALEU(UUR) gene revealed a A to G transition at position 3243, which is a known frequent cause of MELAS syndrome (Figure 2). Restriction digestion with HaeIII of the PCR fragment containing the A3243G mutation revealed an 89% mutational load (Figure 2F). The corresponding kidney tissue was unaffected.
Figure 2.
Mitochondrial DNA analysis of the renal carcinoma with the somatic A3243G mutation. Denaturating HPLC analysis of a PCR fragment (3079–3505) of case 4 at 58°C oven temperature (A–C). The kidney tissue shows a single homoplasmic peak (A), the corresponding renal carcinoma tissue a heteroplasmy over 90% (B) and the mixture of denaturated and reannealed PCR product of kidney and the corresponding tumour tissue resulted in one hetero- and homoduplex peak of the similar height (C). Sequence analysis of the PCR product indicated the wild-type 3243A variant in the unaffected kidney tissue (D), and the 3243G mutation in the corresponding carcinoma tissue (E), as indicated by arrows. Agarose gel analysis of a restriction digestion of a PCR fragment (3118–3332) with HaeIII, which specifically recognizes the 3243G mutation, and two control sites within the PCR fragment yielding two small fragments (F): undigested full-length PCR fragment of 215 base pairs (lane 1); kidney tissue resulting in a 169-base pair fragment (lane 2); carcinoma tissue resulting in a weak 169-base pair fragment of the residual wild-type 3243A variant as well as 72- and 97-base pair fragments, indicating the 3243G mutation (lane 3); 100 bp molecular weight marker (lane 4). R=G+A.
Furthermore, the known MELAS suppresser G12300A mutation (El Meziane et al, 1998) was not present in the tumour sample with the A3243G mutation.
Enzyme activities
Biochemical analysis of OXPHOS and Krebs-cycle enzyme activities of 37 renal carcinoma tissues were reported in our previous study (Meierhofer et al, 2004), which included the 15 patients presented here (Table 3). A total of 14 renal carcinoma tumour tissues, investigated in this study, showed a depletion of the mtDNA content (46%) combined with a decreased activity of complex I (16%), complex II (23%), complex IV (29%), oligomycin-sensitive ATPase activity of complex V (25%) and citrate synthase (72%) compared to matched normal cortex tissues (Table 3) (Meierhofer et al, 2004).
Table 3. Median enzyme activities of case 4 with the somatic A3243G mutation in the tumour tissue compared to 14 matched tissue pairs.
|
Kidney cortex
|
Renal carcinoma tissue
|
|||||
|---|---|---|---|---|---|---|
| Enzyme | Case 4 | Median (n=14) | Range | Case 4 | Median (n=14) | Range |
| CS | 63 | 99 | 60–161 | 362 | 66 | 27–486 |
| CI | 37 | 50 | 11–95 | 8 | 6 | 1–17 |
| CII | 96 | 115 | 31–206 | 132 | 22 | 6–51 |
| CIV | 97 | 181 | 44–278 | 76 | 36 | 21–108 |
| CV | 27 | 48 | 28–125 | 412 | 13 | 0–63 |
Values in U per g protein.
RC=renal carcinoma; CS=citrate synthase; CI=complex I; CII=complex II; CIV=complex IV; CV=complex V.
This is in clear contrast to the tumour tissue with the A3243G mutation, which showed an upregulation of the mtDNA content (170%) and increased enzyme activities of complex II (138%), oligomycin-sensitive ATPase activity of complex V (1530%) and citrate synthase (575%) and a downregulation of the mitochondrial- and nuclear-encoded enzyme activities of complex I (22%) and complex IV (78%) related to the matching control tissue (Table 3).
One tumour tissue displayed only an elevated level of citrate synthase (300%). As expected, no pathogenic mutation was found in the mtDNA of this sample.
Western blot analysis of the VHL tumour suppressor protein
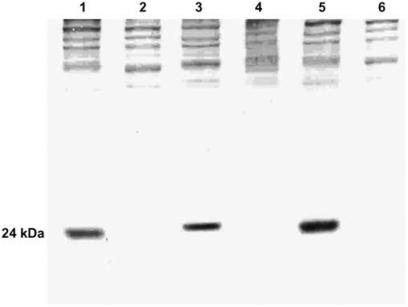
The von Hippel-Lindau tumour suppressor protein (VHL) is frequently absent in renal carcinoma tissues. A recent study has shown that VHL deficiency is one of the factors responsible for downregulation of the biogenesis of OXPHOS complexes in renal carcinoma (Hervouet et al, 2005). To exclude that the altered enzyme activities of the OXPHOS complex in the tumour tissue with the A3243G mutation are based on the presence of the VHL protein, we tested tumour tissues by Western blot analysis for VHL content. In the renal carcinoma tissue harbouring the A3243G mutation as well as in the two other renal carcinoma tissues investigated, lack of VHL protein was observed (Figure 3).
Figure 3.
Western blot analysis of VHL protein content in extracts of three matched tissue pairs. Case 3: kidney control tissue (lane 1) and renal carcinoma tissue (lane 2); case 4: kidney control tissue (lane 3) and renal carcinoma tissue with the somatic A3243G mutation (lane 4); case 5: kidney control tissue (lane 5) and renal carcinoma tissue (lane 6); VHL: 24 kDa.
DISCUSSION
In the present study, we evaluated the influence of mtDNA mutations on the OXPHOS capacity of renal carcinoma tissues. The mtDNA was more frequently mutated in renal carcinoma than in kidney cortex tissues. This is in line with somatic mtDNA mutations reported in a wide variety of human neoplasias (Fliss et al, 2000; Jones et al, 2001; Nagy et al, 2002; Tan et al, 2002). Somatic mutations in coding regions of the mtDNA in tumour tissues are either silent or mostly nonpathogenic polymorphisms. Only a few pathogenic mtDNA mutations have been reported (Brown et al, 1992; Parrella et al, 2001; Lorenc et al, 2003). However, no biochemical analysis has been performed to evaluate the consequences of these mutations in the tumour tissues. Here, we show the first combined genetic and biochemical analysis of a mitochondrial A3243G mutation in a tumour tissue. The renal carcinoma tissue with the somatic A3243G mutation showed a compensatory upregulation of enzyme complexes, which are only nuclear encoded. This is in clear contrast to the coordinated downregulation of all components necessary for mitochondrial energy metabolism, which we found in 92% of all renal carcinoma tissues irrespective of tumour stage and progression (Meierhofer et al, 2004). However, the low activity of the partially mtDNA-encoded complexes of the respiratory chain in the tumour tissue with the somatic A3243G mutation will lead to similar low oxidative capacity as in the other renal cell carcinomas. Accordingly, histology and clinical progression did not differ.
The biochemical consequences of the A3243G mutation in the renal carcinoma tissue are in agreement with the decreased activity of the mitochondrial- and nuclear-encoded complex I and IV reported in muscle biopsy and fibroblasts of patients with MELAS syndrome carrying the A3243G mutation (de Vries et al, 1994; Wilichowski et al, 1998; Pronicki et al, 2002). Our findings indicate that the biochemical changes caused by the pathogenic A3243G mutation are not dependent on tissue type and therefore, it seems to be a general cellular compensatory mechanism.
The discrepancy between the normal enzyme activity of complex II vs the decreased complex I and IV enzyme activities in the sample with the A3243G mutation may lie partly in the fact that the latter complexes are involved in proton pumping, whereas the former is not.
Lack of the VHL protein and somatic alterations in the VHL gene are found in about half of conventional renal carcinomas (Brauch et al, 2000). Hervouet et al (2005) reported that the transfection of the VHL gene in VHL-deficient renal carcinoma cells increases mtDNA and respiratory chain protein contents and permitted the cells to rely on their mitochondrial ATP production to grow in the absence of glucose. Presence of an intact VHL protein in the tumour tissue could have explained the observed upregulation of the enzyme activities in the renal carcinoma tissue with the A3243G mutation. However, the VHL protein was not detectable in our case harboring the A3243G mutation.
In contrast to several other studies reporting mtDNA mutations in cancer tissues, no somatic homoplasmic mutations were detected in the renal samples investigated here. One explanation might be that we used sensitive DHPLC analysis with a detection limit of heteroplasmy of down to 1% and up to 99%, respectively (Meierhofer et al, 2005). Furthermore, contaminations of the tumour tissues with blood and epithelial cells of vessels might contribute to a certain degree of heteroplasmy. However, these contaminations might also have been latent in previous studies. Finally, because of high background in sequence analysis, heteroplasmies of over 80% might appear as homoplasmic mutation and therefore, high-level of heteroplasmy in this study might be equivalent to homoplasmy in other studies (Meierhofer et al, 2005). In accordance, the heteroplasmy in the tumour tissue of case 12 (C8750T) is only visible in DHPLC analysis but not in direct sequence analysis (Figure 1F and H). For the same reason, low-level heteroplasmy might be overlooked by automated sequence analysis.
It is difficult to compare the frequency of mtDNA mutations in different studies analysing mtDNA mutations in distinct cancer types, because most of these studies only analysed parts of the mitochondrial genome, mostly the D-loop. Complete sequence analysis of the mtDNA of 10 primary ovarian carcinomas revealed somatic mtDNA mutations in 60% of the tumour samples (Liu et al, 2001). Another study found that 74% of breast cancer samples had at least one somatic mtDNA mutation (Tan et al, 2002), indicating that the incidence of homoplasmic or high level heteroplasmic somatic mutations in these tumours is higher than in renal cell carcinoma reported here.
Mutations in the D-loop regulatory region might alter the rate of DNA replication by modifying the binding affinity of significant trans-activating factors. In the renal carcinomas presented here, only four out of the 14 somatic mutations affected the D-loop. This cannot explain the decrease of the mtDNA content observed in the majority of renal carcinomas (Simonnet et al, 2002; Meierhofer et al, 2004). In agreement with other studies, either G to A or T to C transitions were observed, which is consistent with the mutagenic spectra of oxidative damage (Lee et al, 2004).
In half of the somatic mutations, the level of heteroplasmy was between 1 and 25%. Phenotypic manifestation of a genetic defect of the mtDNA occurs only if a threshold level is exceeded. Although the phenotypic threshold depends on the type of mutation and tissue, it has been shown that a heteroplasmy at least over 60% is necessary to show effects on the enzyme activity of respiratory chain complexes (Rossignol et al, 2003). Therefore, a primary role of the low heteroplasmic mutations in downregulation of OXPHOS activity and renal carcinogenesis is unlikely. Accumulation of mtDNA mutations in tumours can be explained without selection (Coller et al, 2001). Heteroplasmic mutations might have accumulated in cancer cells by random drift of a pre-existing heteroplasmy (Nekhaeva et al, 2002). This random drift will also explain the observed shift of the degree of heteroplasmy of a mixture of mutated and wild-type mtDNA already detectable in tumour progenitor cells.
In conclusion, the low frequency as well as the low level of heteroplasmy of the somatic mtDNA mutations in renal carcinomas does not indicate a major contribution of these alterations in tumour development. Furthermore, the downregulation of the mitochondrial energy metabolism observed in renal carcinomas cannot be explained by the presence of mtDNA mutations. Even, the case carrying the A3243G mutation exhibits normal tumour histology and low overall oxidative capacity. More likely, a general nuclear-encoded mechanism results in the adaptation of the aerobic energy metabolism in renal carcinoma.
Acknowledgments
This work was supported by the ‘Children's Cancer Foundation Salzburg’ and the ‘Vereinigung zur Foerderung der paediatrischen Forschung und Fortbildung Salzburg’.
References
- Berger A, Mayr JA, Meierhofer D, Foetschl U, Bittner R, Budka H, Grethen C, Huemer M, Kofler B, Sperl W (2003) Severe depletion of mitochondrial DNA in spinal muscular atrophy. Acta Neuropathol 105: 245–251 [DOI] [PubMed] [Google Scholar]
- Brauch H, Weirich G, Brieger J, Glavac D, Rodl H, Eichinger M, Feurer M, Weidt E, Puranakanitstha C, Neuhaus C, Pomer S, Brenner W, Schirmacher P, Storkel S, Rotter M, Masera A, Gugeler N, Decker HJ (2000) VHL alterations in human clear cell renal cell carcinoma: association with advanced tumor stage and a novel hot spot mutation. Cancer Res 60: 1942–1948 [PubMed] [Google Scholar]
- Brown MD, Voljavec AS, Lott MT, Torroni A, Yang CC, Wallace DC (1992) Mitochondrial DNA complex I and III mutations associated with Leber's hereditary optic neuropathy. Genetics 130: 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, George Jr M, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA 76: 1967–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Khrapko K, Bodyak ND, Nekhaeva E, Herrero-Jimenez P, Thilly WG (2001) High frequency of homoplasmic mitochondrial DNA mutations in human tumors can be explained without selection. Nat Genet 28: 147–150 [DOI] [PubMed] [Google Scholar]
- de Vries D, de Wijs I, Ruitenbeek W, Begeer J, Smit P, Bentlage H, van Oost B (1994) Extreme variability of clinical symptoms among sibs in a MELAS family correlated with heteroplasmy for the mitochondrial A3243G mutation. J Neurol Sci 124: 77–82 [DOI] [PubMed] [Google Scholar]
- El Meziane A, Lehtinen SK, Hance N, Nijtmans LG, Dunbar D, Holt IJ, Jacobs HT (1998) A tRNA suppressor mutation in human mitochondria. Nat Genet 18: 350–353 [DOI] [PubMed] [Google Scholar]
- Faure Vigny H, Heddi A, Giraud S, Chautard D, Stepien G (1996) Expression of oxidative phosphorylation genes in renal tumors and tumoral cell lines. Mol Carcinog 16: 165–172 [DOI] [PubMed] [Google Scholar]
- Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, Eleff SM, Jen J, Sidransky D (2000) Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287: 2017–2019 [DOI] [PubMed] [Google Scholar]
- Girald-Rosa W, Vleugels RA, Musiek AC, Sligh JE (2005) High-throughput mitochondrial genome screening method for nonmelanoma skin cancer using multiplexed temperature gradient capillary electrophoresis. Clin Chem 51: 305–311 [DOI] [PubMed] [Google Scholar]
- Hervouet E, Demont J, Pecina P, Vojtiskova A, Houstek J, Simonnet H, Godinot C (2005) A new role for the von Hippel–Lindau tumor suppressor protein: stimulation of mitochondrial oxidative phosphorylation complex biogenesis. Carcinogenesis 26: 531–539 [DOI] [PubMed] [Google Scholar]
- Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A (2001a) Detection of mitochondrial DNA alterations in primary tumors and corresponding serum of colorectal cancer patients. Int J Cancer 94: 429–431 [DOI] [PubMed] [Google Scholar]
- Hibi K, Nakayama H, Yamazaki T, Takase T, Taguchi M, Kasai Y, Ito K, Akiyama S, Nakao A (2001b) Mitochondrial DNA alteration in esophageal cancer. Int J Cancer 92: 319–321 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Nomoto S, Caballero OL, Usadel H, Henrique R, Varzim G, Oliveira J, Lopes C, Fliss MS, Sidransky D (2001) Mitochondrial mutations in early stage prostate cancer and bodily fluids. Oncogene 20: 5195–5198 [DOI] [PubMed] [Google Scholar]
- Jones JB, Song JJ, Hempen PM, Parmigiani G, Hruban RH, Kern SE (2001) Detection of mitochondrial DNA mutations in pancreatic cancer offers a ‘mass’-ive advantage over detection of nuclear DNA mutations. Cancer Res 61: 1299–1304 [PubMed] [Google Scholar]
- Khrapko K, Coller HA, Andre PC, Li XC, Hanekamp JS, Thilly WG (1997) Mitochondrial mutational spectra in human cells and tissues. Proc Natl Acad Sci USA 94: 13798–13803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumimoto H, Yamane Y, Nishimoto Y, Fukami H, Shinoda M, Hatooka S, Ishizaki K (2004) Frequent somatic mutations of mitochondrial DNA in esophageal squamous cell carcinoma. Int J Cancer 108: 228–231 [DOI] [PubMed] [Google Scholar]
- Lee HC, Li SH, Lin JC, Wu CC, Yeh DC, Wei YH (2004) Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res 547: 71–78 [DOI] [PubMed] [Google Scholar]
- Lievre A, Chapusot C, Bouvier AM, Zinzindohoue F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J, Laurent-Puig P (2005) Clinical value of mitochondrial mutations in colorectal cancer. J Clin Oncol 23: 3517–3525 [DOI] [PubMed] [Google Scholar]
- Liu VW, Shi HH, Cheung AN, Chiu PM, Leung TW, Nagley P, Wong LC, Ngan HY (2001) High incidence of somatic mitochondrial DNA mutations in human ovarian carcinomas. Cancer Res 61: 5998–6001 [PubMed] [Google Scholar]
- Lorenc A, Bryk J, Golik P, Kupryjanczyk J, Ostrowski J, Pronicki M, Semczuk A, Szolkowska M, Bartnik E (2003) Homoplasmic MELAS A3243G mtDNA mutation in a colon cancer sample. Mitochondrion 3: 119–124 [DOI] [PubMed] [Google Scholar]
- Maximo V, Soares P, Seruca R, Rocha AS, Castro P, Sobrinho-Simoes M (2001) Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer 32: 136–143 [DOI] [PubMed] [Google Scholar]
- Meierhofer D, Mayr JA, Ebner S, Sperl W, Kofler B (2005) Rapid screening of the entire mitochondrial DNA for low-level heteroplasmic mutations. Mitochondrion 5: 282–296 [DOI] [PubMed] [Google Scholar]
- Meierhofer D, Mayr JA, Foetschl U, Berger A, Fink K, Schmeller N, Hacker GW, Hauser-Kronberger C, Kofler B, Sperl W (2004) Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis 25: 1005–1010 [DOI] [PubMed] [Google Scholar]
- MitoAnalyzer (2000) MitoAnalyzer. Gaithersburg, MD, USA: National Institute of Standards and Technology http://www.cstl.nist.gov/biotech/strbase/mitoanalyzer.html [Google Scholar]
- Nagy A, Wilhelm M, Sukosd F, Ljungberg B, Kovacs G (2002) Somatic mitochondrial DNA mutations in human chromophobe renal cell carcinomas. Genes Chromosomes Cancer 35: 256–260 [DOI] [PubMed] [Google Scholar]
- Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K (2002) Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues. Proc Natl Acad Sci USA 99: 5521–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa M, Nishiguchi S, Shiomi S, Tamori A, Koh N, Takeda T, Kubo S, Hirohashi K, Kinoshita H, Sato E, Inoue M (2001) Somatic mutation of mitochondrial DNA in cancerous and noncancerous liver tissue in individuals with hepatocellular carcinoma. Cancer Res 61: 1843–1845 [PubMed] [Google Scholar]
- Parrella P, Xiao Y, Fliss M, Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E, Cuomo C, Cohen D, Pandit S, Spencer M, Rabitti C, Fazio VM, Sidransky D (2001) Detection of mitochondrial DNA mutations in primary breast cancer and fine-needle aspirates. Cancer Res 61: 7623–7626 [PubMed] [Google Scholar]
- Pejovic T, Ladner D, Intengan M, Zheng K, Fairchild T, Dillon D, Easley S, Marchetti D, Schwartz P, Lele S, Costa J, Odunsi K (2004) Somatic D-loop mitochondrial DNA mutations are frequent in uterine serous carcinoma. Eur J Cancer 40: 2519–2524 [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, Marshall FF, Wallace DC (2005) mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA 102: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettepher CC, LeDoux SP, Bohr VA, Wilson GL (1991) Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem 266: 3113–3117 [PubMed] [Google Scholar]
- Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, Markowitz SD, Trush MA, Kinzler KW, Vogelstein B (1998) Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet 20: 291–293 [DOI] [PubMed] [Google Scholar]
- Pronicki M, Sykut-Cegielska J, Mierzewska H, Tonska K, Karczmarewicz E, Iwanicka K, Bartnik E, Pronicka E (2002) Diversity of clinical symptoms in A3243G mitochondrial DNA mutation (MELAS syndrome mutation). Med Sci Monit 8: CR767–CR773 [PubMed] [Google Scholar]
- Richard SM, Bailliet G, Paez GL, Bianchi MS, Peltomaki P, Bianchi NO (2000) Nuclear and mitochondrial genome instability in human breast cancer. Cancer Res 60: 4231–4237 [PubMed] [Google Scholar]
- Rossignol R, Faustin B, Rocher C, Malgat M, Mazat JP, Letellier T (2003) Mitochondrial threshold effects. Biochem J 370: 751–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin P, Chretien D, Bourgeron T, Gerard B, Rotig A, Saudubray JM, Munnich A (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin Chim Acta 228: 35–51 [DOI] [PubMed] [Google Scholar]
- Shiao YH, Resau JH, Nagashima K, Anderson LM, Ramakrishna G (2000) The von Hippel–Lindau tumor suppressor targets to mitochondria. Cancer Res 60: 2816–2819 [PubMed] [Google Scholar]
- Shidara Y, Yamagata K, Kanamori T, Nakano K, Kwong JQ, Manfredi G, Oda H, Ohta S (2005) Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis. Cancer Res 65: 1655–1663 [DOI] [PubMed] [Google Scholar]
- Silvestri G, Rana M, DiMuzio A, Uncini A, Tonali P, Servidei S (1998) A late-onset mitochondrial myopathy is associated with a novel mitochondrial DNA (mtDNA) point mutation in the tRNA(Trp) gene. Neuromuscul Disord 8: 291–295 [DOI] [PubMed] [Google Scholar]
- Simonnet H, Alazard N, Pfeiffer K, Gallou C, Beroud C, Demont J, Bouvier R, Schagger H, Godinot C (2002) Low mitochondrial respiratory chain content correlates with tumor aggressiveness in renal cell carcinoma. Carcinogenesis 23: 759–768 [DOI] [PubMed] [Google Scholar]
- Storkel S, Eble JN, Adlakha K, Amin M, Blute ML, Bostwick DG, Darson M, Delahunt B, Iczkowski K (1997) Classification of renal cell carcinoma: Workgroup No. 1. Union Internationale Contre le Cancer (UICC) and the American Joint Committee on Cancer (AJCC). Cancer 80: 987–989 [DOI] [PubMed] [Google Scholar]
- Tan DJ, Bai RK, Wong LJ (2002) Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res 62: 972–976 [PubMed] [Google Scholar]
- Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE (2003) Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 3: 1620–1632 [DOI] [PubMed] [Google Scholar]
- Uziel G, Moroni I, Lamantea E, Fratta GM, Ciceri E, Carrara F, Zeviani M (1997) Mitochondrial disease associated with the T8993G mutation of the mitochondrial ATPase 6 gene: a clinical, biochemical, and molecular study in six families. J Neurol Neurosurg Psychiatry 63: 16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch BJ, de Coo IF, Hendrickx AT, Busch HF, de Jong G, Scholte HR, Smeets HJ (2004) Increased risk for cardiorespiratory failure associated with the A3302G mutation in the mitochondrial DNA encoded tRNALeu(UUR) gene. Neuromuscul Disord 14: 683–688 [DOI] [PubMed] [Google Scholar]
- Warburg O (1956) On the origin of cancer cells. Science 123: 309–314 [DOI] [PubMed] [Google Scholar]
- Wilichowski E, Korenke GC, Ruitenbeek W, De Meirleir L, Hagendorff A, Janssen AJ, Lissens W, Hanefeld F (1998) Pyruvate dehydrogenase complex deficiency and altered respiratory chain function in a patient with Kearns-Sayre/MELAS overlap syndrome and A3243G mtDNA mutation. J Neurol Sci 157: 206–213 [DOI] [PubMed] [Google Scholar]
- Wu CW, Yin PH, Hung WY, Li AF, Li SH, Chi CW, Wei YH, Lee HC (2005) Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer 44: 19–28 [DOI] [PubMed] [Google Scholar]
- Yeh JJ, Lunetta KL, van Orsouw NJ, Moore Jr FD, Mutter GL, Vijg J, Dahia PL, Eng C (2000) Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene 19: 2060–2066 [DOI] [PubMed] [Google Scholar]
- Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER (2005) Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis 26: 145–152 [DOI] [PubMed] [Google Scholar]