Abstract
In order to elucidate the biological variance between normal ovarian surface epithelial (NOSE) and epithelial ovarian cancer (EOC) cells, and to build a molecular classifier to discover new markers distinguishing these cells, we analysed gene expression patterns of 65 primary cultures of these tissues by oligonucleotide microarray. Unsupervised clustering highlights three subgroups of tumours: low malignant potential tumours, invasive solid tumours and tumour cells derived from ascites. We selected 18 genes with expression profiles that enable the distinction of NOSE from these three groups of EOC with 92% accuracy. Validation using an independent published data set derived from tissues or primary cultures confirmed a high accuracy (87–96%). The distinctive expression pattern of a subset of genes was validated by quantitative reverse transcription–PCR. An ovarian-specific tissue array representing tissues from NOSE and EOC samples of various subtypes and grades was used to further assess the protein expression patterns of two differentially expressed genes (Msln and BMP-2) by immunohistochemistry. This study highlights the relevance of using primary cultures of epithelial ovarian cells as a model system for gene profiling studies and demonstrates that the statistical analysis of gene expression profiling is a useful approach for selecting novel molecular tumour markers.
Keywords: microarray, ovarian cancer, molecular marker, mesothelin, BMP-2
Epithelial ovarian cancer (EOC), a complex disease, is the second most common gynaecological cancer, and accounts for nearly half of the deaths associated with gynaecological pelvic malignancies. Largely asymptomatic, over 70% of patients with ovarian cancer are already at an advanced stage of the disease at initial diagnosis. Considering the morphological, anatomical and clinical differences between tumours, three main classification parameters have emerged, which are based on histological subtype (serous, endometrioid, clear cells, mucinous, Brenner), disease stage (I–IV) and tumour grade (0–3). However, early detection of ovarian cancer is rare, and screening program in the general population have been unsuccessful. Recent studies have focused on gene expression patterns observed for multiple tumours to identify the molecular events involved in the development of cancer in order to uncover diagnostic and prognostic markers as well as new therapeutic targets. Promising results have been reported in several cancers, including bladder, colon and breast cancers (Neibergs et al, 2002; van’t Veer et al, 2002; Dyrskjot et al, 2003), of gene expression profiling as a new means for identifying diagnostic and prognostic targets. While this approach has been applied to ovarian cancer (Adib et al, 2004; Donninger et al, 2004; Hibbs et al, 2004; Lancaster et al, 2004; Lee et al, 2004; Lu et al, 2004; Santin et al, 2004; Warrenfeltz et al, 2004; Zhang et al, 2005), resulting in the identification of several hundred genes differentially expressed between NOSE (normal ovarian surface epithelia) and EOC, less than 10% of these genes were identified in common by at least two reports (Le Page et al, 2004). Although sample size, methods of evaluation and expression platforms vary and could account for differences in gene expression profiles, additional independent studies are required for cross-comparison to identify reliable gene markers that vary in expression.
Gene profiling by microarray studies can also be applied to tumour classification, identification of tumour subtypes, or used to predict disease outcome. Expression-based classification schemes have been applied to distinguish ovarian cancers from other tumour types (Golub et al, 1999; Ross et al, 2000; Ramaswamy et al, 2001), but rarely to distinguish normal samples from ovarian cancers. A recent study classified pooled normal epithelial samples from ovarian tumours based on the gene expression of CLDN3 and VEGF (Lu et al, 2004). The choice of model system to study molecular profiling is a critical factor that may account for variations in gene expression patterns. As normal epithelial cells are generally quiescent, differential gene expression observed between tissues of NOSE and EOC may reflect differences not necessarily related to oncogenic transformation. In ovarian cancer, primary cultures derived from NOSE and tumour tissue have been described (Lounis et al, 1994). In the case of EOC tissues, primary cultures have the advantage of being relatively free of non-transformed cell population. NOSE cultures, derived by scraping the surface of the ovary, are highly homogenous and provide conditions in which normal cells replicate over a limited number of generations. In addition, a recent study comparing candidate genes able to stratify low- from high-malignancy tumours (identified in primary cultures and tumour tissues) showed that candidate genes identified in primary cultures were also able to reclassify samples derived from tumour tissues (Ouellet et al, 2005). Based on these advantages, we studied primary cultures enriched over several passages. Using 65 independent samples, we identified gene expression patterns by Affymetrix oligonucleotide microarray analyses, which enabled the stratification of NOSE from EOC and was further supported by tissue array analyses.
MATERIALS AND METHODS
Patients, cell culture and clinical material
Tissue samples and sera were obtained with informed consent from participants. Tumour samples were collected from surgeries performed at the Centre Hospitalier de l’Université de Montréal (CHUM). Histopathology, grade and stage of tumours were assigned according to the International Federation of Gynecology and Obstetrics (FIGO) criteria. Normal tissues were obtained from tumour-free participants that have undergone oopherectomies. Primary cell cultures from NOSE and EOC samples were established as described (Kruk et al, 1990; Lounis et al, 1994) and used for microarray analysis. Cells in primary culture were maintained in OSE media consisting of 50 : 50 medium 199:105 (Sigma, St Louise, MO, USA) supplemented with 10% fetal bovine serum, 2.5 μg ml−1 amphotericin B and 50 μg ml−1 gentamicin (Kruk et al, 1990). The samples used for microarray, reverse transcription (RT)–PCR and tissue array immunohistochemistry (IHC) studies are presented in Table 1.
Table 1. Specimen and clinical data of ovarian cancer patients used in each experiments.
|
Grade
|
Stage
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Experiments | Samples | LMP | 1 | 2 | 3 | Unknown | Low (1–2) | High (3–4) | Solid tumours | Ascites |
| Training set (n=65) | Normal n=11 | |||||||||
| Serous n=40 | 6 | 1 | 8 | 25 | 4 | 36 | 23 | 17 | ||
| Endometrioid n=9 | 3 | 6 | 3 | 6 | 5 | 5 | ||||
| Clear cell n=2 | 2 | 1 | 1 | 0 | 2 | |||||
| Mixed n=1 | 1 | 1 | 1 | 0 | ||||||
| Mucinous=1 | 1 | 1 | ||||||||
| Undifferentiated n=1 | 3 | 0 | 1 | |||||||
| Total tumours n=54 | 7 | 1 | 11 | 37 | 1 | 10 | 44 | 29 | 25 | |
| Test set primary culture (n=23) | Normal and benign n=6 | |||||||||
| Serous n=11 | 3 | 3 | 5 | 3 | 7 | 1 | 9 | |||
| Endometrioid n=1 | 1 | 1 | 1 | 0 | ||||||
| Clear cell n=1 | 1 | 1 | 0 | 1 | ||||||
| Mixed n=3 | 3 | 3 | 1 | 2 | ||||||
| Undifferentiated n=1 | 1 | 1 | 1 | 0 | ||||||
| Total tumours n=17 | 3 | 3 | 11 | 3 | 13 | 4 | 12 | |||
| Test set tissue (n=137)a | Normal and benign n=5 | NS | NS | 5 | 0 | |||||
| Serous n=81 | 6 | 3 | 21 | 41 | 10 | NS | NS | 81 | 0 | |
| Endometrioid n=33 | 10 | 10 | 13 | NS | NS | 33 | 0 | |||
| Clear cell n=8 | 8 | NS | NS | 8 | 0 | |||||
| Mucinous n=10 | 7 | 3 | NS | NS | 10 | 0 | ||||
| Total tumours n=132 | 6 | 20 | 34 | 62 | 10 | NS | NS | 137 | 0 | |
| Tissue array (n=89) | Normal n=20 | |||||||||
| Serous n=21 | 4 | 5 | 5 | 7 | 1 | 8 | 13 | 21 | 0 | |
| Endometrioid n=27 | 13 | 7 | 5 | 2 | 17 | 10 | 27 | 0 | ||
| Clear cell n=17 | 5 | 9 | 3 | 11 | 6 | 17 | 0 | |||
| Mixed n=4 | 3 | 1 | 2 | 2 | 4 | 0 | ||||
| Total tumours n=69 | 4 | 18 | 17 | 24 | 6 | 38 | 31 | 69 | 0 | |
LMP=low malignant potential.
From publicly available data sets (Welsh et al, 2001; Schwartz et al, 2002; Ouellet et al, 2005). ns: nonspecified.
RNA preparation and microarray
Total RNA was extracted with TRIzol™ reagent (Gibco/BRL, Life Technologies Inc., Grand Island, NY, USA). RNA was extracted directly from cells grown to 80% confluency. RNA quality was monitored by agarose gel electrophoresis, and independently by the 2100 bioanalyzer using the RNA 6000 Nano LabChip kit (Agilent Technologies, Germany). Biotinylated hybridisation target was prepared from total RNA as described (Tamayo et al, 1999). Affymetrix HuFL arrays were used to hybridise label targets, and gene expression levels were calculated for each EST from the scanned image by the Affymetrix GeneChip MAS4 software. This gene chip contains 6800 probe sets representing known genes and ESTs (Affymetrix.com). Microarray experiments were performed at the McGill University and Genome Québec Innovation Centre. The detailed protocol is available at www.genomequebec.mcgill.ca/center.php. The raw data of each experiment, including data used for training and test sets, were normalised according to the mean of the global intensity expression values adjusted to 100 units. After normalisation, we considered all values below 20 as technical noise and rescaled these values to 20. Next, we removed for further analysis all expression values for a given probe set with a poor reliability score or ‘A’ call (ambiguous signal) that occurred across all samples. Using this filtering approach, the expression values of 4888 probe sets were examined for patterns of gene expression. Values were then converted into log10 for further analysis. Both the raw and normalised data sets are available at http://www.genomequebec.mcgill.ca/ovarian/.
Clustering
We carried out hierarchical clustering analysis using GeneSpring™ software (Silicone Genetics) on the filtered data set. We used the distance branch of 0.1 with a Pearson correlation as a similarity metric.
Supervised class comparison
Three statistical tests were used to identify classifiers. The signal-to-noise metric Sx=(μ1−μ2)/(σ1+σ2) was applied, where μ represents the mean and σ the standard deviation of either class 1 or 2 (Golub et al, 1999). Here, class 1 represents NOSE samples and class 2 represents tumour samples. Only genes with ∣Sx∣⩾0.05 were selected. Classifiers were also identified using the significance analysis of microarray (SAM) software described (Tusher et al, 2001) and available at http://www-stat-class.standford.edu/SAM/SAMServlet. One thousand permutations were applied in an unpaired filtered data set. The threshold was chosen according to a determined false discovery rate (FRD<5%). A non-parametric test was also performed using the Mann–Whitney U-test included in the GeneSpring™ software, including an FDR correction (P-value <0.05 or P<0.15) (Silicone Genetics).
Tumour classification and prediction
To classify samples, we used a k-nearest neighbour algorithm included in the GeneSpring™ software (Silicon Genetics). Leave-one-out cross-validation approach was used to evaluate the predictors in the training set. A class prediction test was used to predict the independent test samples based on classifiers defined from the training set. Each sample from the test set is classified by finding the k-nearest neighbouring training samples. The neighbour number was n−1, where n is the smallest number of samples in one group. The decision cutoff was P<0.2. All sample sets (training and test sets) were evaluated with the same algorithm.
Quantitative PCR
As previously described (Ouellet et al, 2005), RNA was linearly amplified by Alethia Biotherapeutics (Montreal, QC, Canada). The RAMP RNA produced is (+) sense. The cDNA synthesis was performed according to the protocol of the SuperScript™ First-Strand Synthesis System for RT–PCR (Invitrogen Life Technologies) with a starting amount of 2 μg of RNA; the RT was performed with random hexamers. The condition of the PCR reaction (temperature, specificity) was defined by conventional PCR. Positive and negative controls were introduced in all experiments. Quantitative PCR (Q-PCR) was performed using Rotor-gene 3000 Real-Time Centrifugal DNA Amplification System (Corbet Tumor Tissues Research, NSW, Australia). We used the Quantitect™ SYBR Green PCR (Qiagen Inc., ON, Canada) reaction mixture according to the manufacturer's instructions. Serial dilutions were performed to generate a standard curve for each gene tested in order to define the efficiency of the Q-PCR reaction, and a melt curve was constructed to confirm the specificity of the reaction. Experiments were performed in duplicate. Control RNA (Erk) was chosen based on stable expression in 69 ovarian samples containing normal and tumour types as previously described (Ouellet et al, 2005). Primers are described in Supplementary Table 2. We used the Pfaffl analysis method to measure the relative quantity of gene expression (Pfaffl, 2001). Statistical analyses were performed using the Student's t-test.
Tissue array and IHC
The following monoclonal antibodies were used in IHC: anti-ATPaseβ1, anti-BMP-2, anti-TNFR1 (Santa-Cruz Biotechnology, CA, USA) and anti-mesothelin (Msln) (Biogenex, San Roman, CA, USA). A tissue array containing 94 cores of ovarian epithelial tissues (Table 1) was amassed and used for IHC studies. Briefly, the tissue array was heated at 60°C for 30 min, de-paraffinised in toluene and rehydrated in a gradient of ethanol. To unmask antigen, the slides were submerged in 90°C citrate buffer (0.01 M citric acid+500 μl Tween 20 l−1 adjusted to pH 6.0) (JT Baker, Philipsburg, NJ, USA) for 15 min. The tissue was blocked with a protein-blocking serum-free reagent (DakoCytomation Inc., Mississauga, ON, Canada) and incubated with different antibodies overnight at 4°C in a humid chamber. The optimal concentration for each primary antibody was determined by serial dilutions. Subsequently, endogenous peroxidase activity was quenched by treatment with 3% H2O2. The array was then incubated with a secondary biotinylated antibody (DakoCytomation Inc., Mississauga, ON, Canada) for 10 min, followed by incubation with a streptavidin–peroxidase complex (Dako Diagnostics Canada Inc., Mississauga, ON, Canada) for 10 min at room temperature. Reaction products were developed using diaminobenzidine (brown stain) containing 0.3% H2O2 as a substrate for peroxidase, and nuclei were counterstained with diluted haematoxylin (blue stain). Epithelial zones were scored according to the intensity of staining (0 for absence, 1 for very weak, 2 for weak, 3 for moderate and 4 for high intensity). Each array was independently analysed in a blinded study by two independent observers. Statistical analyses were performed using the Mann–Whitney U-test.
RESULTS
Comparative gene profiling of normal epithelial cells (NOSE) and epithelial ovarian tumours (EOC)
To identify markers that may be associated with the progression towards a malignant phenotype, we used oligo-microarrays to analyse gene expression profiles of normal or tumoral primary cultures of ovarian epithelial cells. Primary cultures were derived from specimens selected on the basis of the disease course before treatment of the patient (e.g. no chemotherapy). Primary cultures from 54 malignant tumours and 11 NOSE samples were analysed by microarray. The samples represent as much as possible the diversity of EOC in terms of grade and histopathology (Table 1); Brenner tumours were not represented in our set.
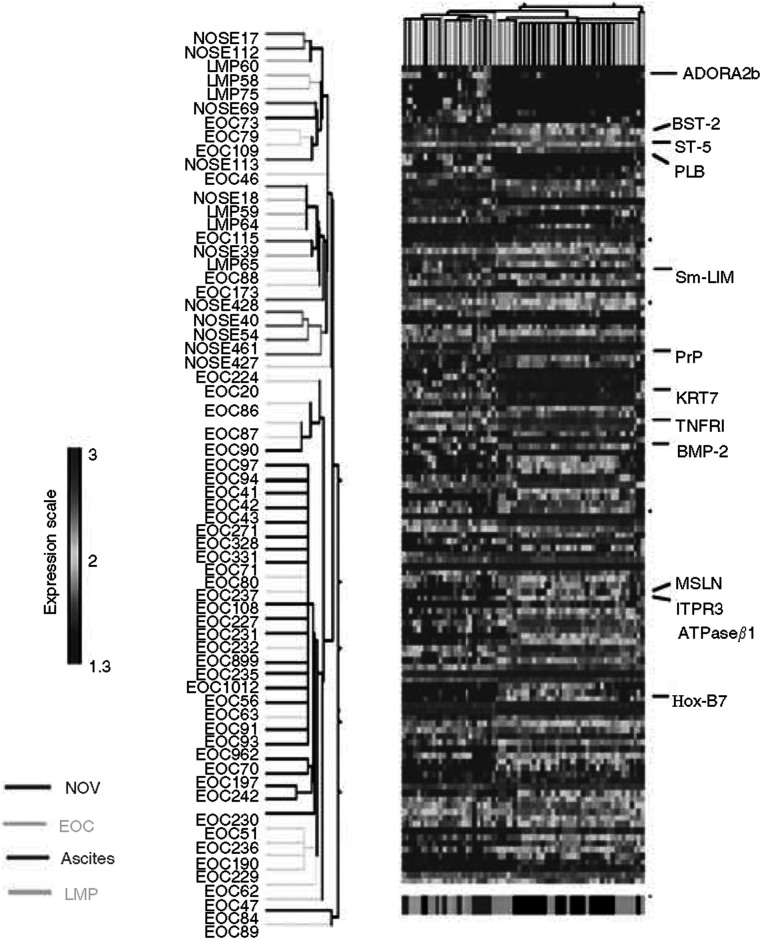
To gain insight into genes whose expression is associated with ovarian malignant transformation, we analysed the expression array data derived from HuGeneFL 6800 Affymetrix GeneChip® using three different supervised classification methods. The group of NOSE samples was compared to the 54 EOC samples. A total of 505 genes were selected by at least one algorithm (data not shown), where 126 ESTs were identified in common by all three algorithms that represent genes that were significantly differentially expressed when NOSE samples were compared with EOC samples (Figure 1 and Supplementary Table 1). About 25% of these genes have previously been shown to be differentially expressed in ovarian cancer cells relative to normal ovarian cells (partially shown in Table 2) (Le Page et al, 2004). The 126 genes are associated with known biological functions such as angiogenesis, growth and proliferation, signalling, cell adhesion and metastases, and are potential oncogenes and tumour suppressors (Table 2). The expression profile of these 126 genes, subjected to a two-way hierarchical cluster analysis (Figure 1), identified several characteristic profiles that differed among the groups analysed. The NOSE samples clustered together with the low malignant potential (LMP) tumours and three grade 2 tumours, forming a group considered as low-grade samples. The majority of tumours are grouped in a second cluster independently of histopathological type. We also noticed that ascites show a more homogenous profile in comparison to the solid invasive tumours and, clustered in a sub-branch. The unsupervised clustering analysis showed that these 126 genes were not able to discriminate NOSE samples from LMP samples in contrast to invasive EOC samples. This suggests that LMP tumours should be analysed separately from the invasive tumours to identify specific gene markers for low-grade disease.
Figure 1.
Hierarchical clustering of gene analysis expression of 11 normal ovarian epithelia (NOSE) and 55 epithelial ovarian tumours (EOC) from primary culture. Sample clusters based on 126 genes differentially expressed in tumour samples vs normal epithelia. Clustering was carried out based on the genes retrieved by class comparison (P-value <0.05, FDR<0.05, ∣Sx∣>0.50). Genes were selected by three statistic algorithms (see Materials and Methods). Each row represents a gene and each column represents a sample. Identity of each sample in the clusters is shown on the left of the figure. Colour intensity represents level of gene expression transformed in log 10. LMP=low malignant potential tumours; Msln refers to the protein; MSLN refers to the gene.
Table 2. Biological function of genes deregulated in EOC.
| Gene ID | Unigene_ID | Title | Gene | Regulation in EOC | P a | Reported in other studies in relation to ovarian cancer | |
|---|---|---|---|---|---|---|---|
| Cell cycle/growth | K01911_at | Hs.1832 | Neuropeptide Y | NPY | Up | 0.0035 | No |
| D21878_at | Hs.169998 | Bone marrow stromal cell antigen 1 | BST1 | Up | 2E−05 | No | |
| U72066_at | Hs.29287 | Retinoblastoma-binding protein 8 | RBBP8 | Up | 2E−05 | No | |
| Oncogenes or tumour suppressors | X77548_at | Hs.99908 | Nuclear receptor coactivator 4 | NCOA4 | Up | 0.0005 | No |
| J04102_at | Hs.85146 | v-ets avian erythroblastosis virus E26 oncogene homolog 2 | ETS2 | Up | 0.001 | Ni | |
| L20861_at | Hs.152213 | Wingless-type MMTV integration site family, member 5A | WNT5A | Up | 3E−05 | Yes | |
| X16662 | Hs.87268 | Annexin A8 | ANXA8 | Up | 5E−06 | No | |
| U61262_at | Hs.90408 | Neogenin (chicken) homolog 1, netrin receptor | NEO1 | Up | 2E−06 | Yes | |
| U15131_at | Hs.79265 | Suppression of tumorigenicity 5 | ST5 | Down | 2E−05 | No | |
| Angiogenesis | M34539 | Hs.752 | FK506-binding protein 1A (12kD) | FKBP1A/FKPB12 | Up | 7E−05 | No |
| M30257 | Hs.109225 | Vascular cell adhesion molecule 1 | VCAM1 | Up | 5E−05 | Yes | |
| D49950_at | Hs.83077 | Interleukin 18 (interferon-gamma-inducing factor) | IL18 | Up | 2E−05 | Yes | |
| M31551_s_at | M31551 | All_M31551 576-1134, Human urokinase inhibitor (PAI-2) gene | PAI-2 | Up | 0.0002 | Yes | |
| J04513_at | J04513 | Human basic fibroblast growth factor (bFGF/FGF2) | bFGF | Up | 0.0002 | Yes | |
| X72012_at | Hs.76753 | Endoglin (Osler–Rendu–Weber syndrome 1) | ENG | Down | 0.0002 | Yes | |
| M22960_at | Hs.73853 | Bone morphogenetic protein 2 | BMP-2 | Up | Yes | ||
| Adhesion | D13666 | Osteoblast specific factor 2 (fasciclin I-like) | OSF-2 | Up | 0.0002 | Yes | |
| D84424 | Hs.57697 | Hyaluronan synthase 1 | HAS1 | Up | 0.0006 | No | |
| M28882 | Hs.211579 | Melanoma adhesion molecule | MCAM | Down | 2.7E−6 | No | |
| M29277 | Hs.211579 | Melanoma adhesion molecule | MCAM | Down | 8.54E−6 | No} | |
| M59911 | Hs.265829 | Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | ITGA3 | Up | 5E−06 | No | |
| U41767 | Hs.92208 | A disintegrin and metalloproteinase domain 15 (metargidin) | ADAM15 | Up | 0.0042 | Yes | |
| Y00097_s_at | Hs.118796 | Annexin A6 | ANXA6 | Down | 1E−05 | No | |
| Z26653 | Hs.75279 | Laminin, alpha 2 (merosin, congenital muscular dystrophy) | LAMA2 | Down | 4E−05 | Yes | |
| D49950 | Hs.83077 | Interleukin 18 (interferon-gamma-inducing factor) | IL18 | Up | 2E−05 | Yes | |
| M30257_s_at | Hs.109225 | Vascular cell adhesion molecule 1 | VCAM1 | Up | 5E−05 | Yes | |
| U40282_at | Hs.6196 | Integrin-linked kinase | ILK | Down | 1E−05 | Yes | |
| Metastasis | D21337_at | Hs.408 | Collagen, type IV, alpha 6 | COL4A6 | Up | 0.001 | Yes |
| M90657_at | Hs.3337 | Transmembrane 4 superfamily member 1 | TM4SF1 | Up | 0.0009 | No | |
| M22489_at | Hs.73853 | Bone morphogenetic protein 2 | BMP2 | Up | 0.0006 | Yes | |
| L20861 | Hs.152213 | Wingless-type MMTV integration site family, member 5A | WNT5A | Up | 3E−05 | Yes | |
| Transduction signal/transcription factor | D25538 | Hs.172199 | Adenylate cyclase 7 | ADCY7 | Down | 2E−05 | No |
| L07597 | Hs.149957 | Ribosomal protein S6 kinase, 90kD, polypeptide 1 | RPS6KA1 | Up | 3E−05 | No | |
| M64497 | Hs.288869 | Nuclear receptor subfamily 2, group F, member 2 | NR2F2 | Down | 0.0004 | No | |
| U24576 | Hs.3844 | LIM domain only 4 | LMO4 | Up | 1E−06 | No | |
| U28833 | Hs.86724 | Down syndrome critical region gene 1 | DSCR1 | Up | 3E−06 | No | |
| X68487 | Hs.45743 | Adenosine A2b receptor | ADORA2B | Down | 1E−06 | No | |
| J03161 | Hs.155321 | Serum response factor (c-fos serum response transcription factor) | SRF | Down | 0.001 | No | |
| M62402 | Hs.274313 | Insulin-like growth factor binding protein 6 | IGFBP6 | Down | 2E−05 | Yes | |
| M62403 | Hs.1516 | Insulin-like growth factor-binding protein 4 | IGFBP4 | Up | 0.0004 | Yes | |
| L20861 | Hs.120 | Wingless-type MMTV integration site family, member 5A | WNT5A | Up | 3E−05 | Yes | |
| M34539 | Hs.752 | FK506-binding protein 1A (12kD) | FKBP1A/FKPB12 | Up | 7E−05 | No |
EOC=epithelial ovarian cancer.
Mann–Whitney test.
Selection of genes distinguishing subclasses of ovarian tumours
Based on the results of our initial analysis, we subsequently used a supervised approach to select genes that were able to differentiate the major groups of tumours (LMP, ascites and solid invasive tumours) that clustered in the unsupervised analysis. For this purpose, the three groups of tumours were individually compared to the NOSE samples. Ten genes were differentially expressed in LMP tumour cells (Figure 2A), 16 genes were differentiated in solid invasive tumours (Figure 2B) and 270 genes were differentially expressed in ascites tumours (Figure 2C). Among these 270 genes were the 16 differentially expressed genes identified in the solid invasive tumour group of invasive tumours.
Figure 2.
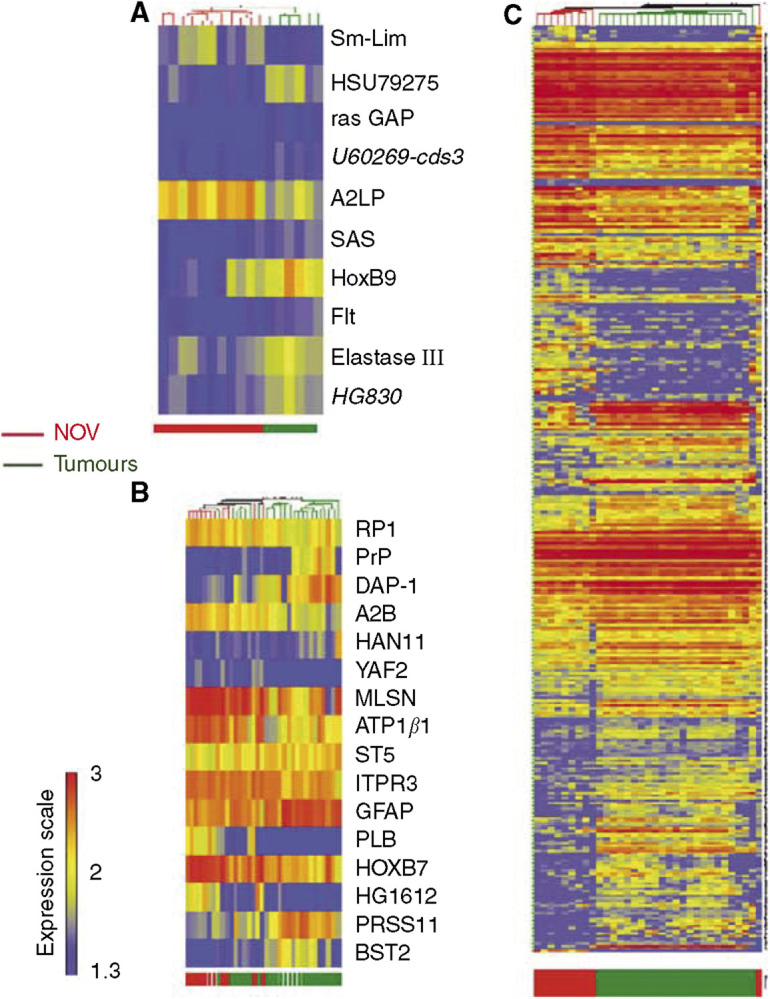
(A) Hierarchical clustering of gene analysis expression of 11 normal ovarian epithelia and six EOC of low malignant potential. Sample clusters and gene profile based on 11 genes differentially expressed in LMP samples. Genes were identified using U algorithm (P<0.15). (B) Hierarchical clustering of gene analysis expression of 11 ovarian epithelia and 24 solid invasive ovarian tumours. Supervised sample clusters and gene profile based on 16 genes differentially expressed in malignant tumour samples are shown. Genes were identified using U algorithm (P<0.05). (C) Hierarchical clustering of gene analysis expression of 11 ovarian epithelia and 23 ovarian samples from malignant ascites. Supervised sample clusters and gene profile based on 270 genes differentially expressed ascites are shown. Genes were identified using U algorithm (P<0.05). Each row represents a gene and each column represents a sample. Colour intensity represents level of gene expression transformed in log 10 corresponding to the expression scale shown on the left side of the figure. NOSE=normal ovarian epithelia; EOC=epithelial ovarian cancer. Colour bar at the bottom of the gene expression matrice shows the class of sample.
As there are no common genes found in the independent analyses of LMP, ascites and invasive tumour groups that distinguished them from NOSE samples, we tested as a classified model the 10 genes differentially expressed in LMP combined with the 16 differentially expressed genes found in common in the analyses of ascites and invasive solid tumours. As an objective prediction of low- and high-malignant ovarian tumours using a limited set of genes could be appropriate for clinical use, we selected gene classifiers showing at least a 1.5-fold gene expression difference between NOSE samples and tumour samples. Using this approach, we reduced the original set of 26 markers to a subset of 18 markers that were the most predictive (Table 3). Each of the 65 tumours was reclassified according to the expression pattern of the 18 gene classifiers. This classifier predicted correctly 60 out of 65 samples (92%) (Table 4). All normal and ascites samples were correctly classified, and only three solid tumours (1.5%) were incorrectly classified as NOSE. This classifier showed greater accuracy in comparison to the large set of 126 genes selected in Figure 1B (Table 4). The incorrectly classified samples included one grade B (LMP) and one grade 2 serous tumours as well as one unknown grade tumour. Interestingly, this classifier allowed a lower ratio of errors/correctly classified (r=0.05) than the set of 126 genes. To validate the classifier, an additional independent test set of 23 primary culture samples and 137 arrays of surgical specimens from publicly available data sets derived from Affymetrix HuFL GeneChip platform (Welsh et al, 2001; Schwartz et al, 2002; Ouellet et al, 2005) were tested. The set of 23 primary culture samples contained two LMP serous ascites. All these samples were normalised the same way as our training set. The algorithm used for class prediction was the same as previously used (k-nearest neighbour). Instead of a cross-validation scheme, these new samples were predicted using a class prediction scheme. The tumour potential was predicted, and resulted in 87% accuracy for the primary culture sample set and 96% accuracy for the tissue sample set (Table 4).
Table 3. Description of 18 genes forming the ovarian tumour classifier.
| Probe set HUFL | Unigene | Name | Symbol | Regulation in ovarian tumours | Ratio | Pa | Classifier of malignancy |
|---|---|---|---|---|---|---|---|
| X94232_at | Hs.78335 | Microtubule-associated protein 2, RP family | RP1/MAPRE2 | Down | 0.6 | 2E−04 | Invasive |
| X83416_s_at | PrP gene, exon 2 | PRP | Up | 2.1 | 1E−04 | Invasive | |
| X68487_at | Hs.45743 | A2b adenosine receptor. | ADORA2B | Down | 0.46 | 2E−05 | Invasive |
| U40434_at | Hs.155981 | Mesothelin or CAK1 antigen precursor mRNA | MSLN | Up | 2.5 | 2E−04 | Invasive |
| U16799_s_at | Hs.78629 | Na,K-ATPase beta-1 subunit mRNA | ATPaseβ1 | Up | 4.5 | 2E−05 | Invasive |
| U15131_at | Hs.79265 | p126 (ST5) mRNA, complete cds. | ST-5/HTS1 | Down | 0.6 | 2E−04 | Invasive |
| U01062_at | Hs.77515 | type 3 inositol 1,4,5-trisphosphate receptor (ITPR3) | ITPR3 | Up | 2.7 | 2E−04 | Invasive |
| M63603_at | Hs.85050 | Phospholamban mRNA, complete cds. | PLN | Down | 0.3 | 4E−05 | Invasive |
| M16937_at | Hs.819 | Homeobox c1 protein; homeobox B7 | HoxB7 | Up | 2.5 | 1E−05 | Invasive |
| HG1612-HT1612_at | MARCKS | Down | 0.4 | 5E−05 | Invasive | ||
| D28137_at | Hs.118110 | BST-2 | BST-2 | Down | 0.5 | 2E−04 | Invasive |
| U79275_at | Hs.27414 | Human clone 23947 mRNA | HSU79275 | Up | 2 | 0.149 | LMP |
| U70671_at | Hs.43509 | A2RP; Human ataxin-2 related protein mRNA, partial cds. | A2RP | Down | 0.5 | 0.149 | LMP |
| U60269_cds3_at | U606269 | Up | 1.5 | 0.149 | LMP | ||
| U46006_s_at | Hs.10526 | Smooth muscle lim protein | SmLIM | Down | 0.6 | 0.149 | LMP |
| U07664_at | HB9 homeobox gene, exons 2 and 3 and complete cds. | HoxB9 | Up | 2.5 | 0.114 | LMP | |
| M18700_s_at | Elastase III A gene, exon 8 | ELIII | Up | 1.9 | 0.093 | LMP | |
| HG830-HT830_at | HG830-Ht830 | Up | 1.7 | 0.093 | LMP |
Gene classifier list of 18 genes discriminating low malignant potential (LMP) tumours and invasive tumours from normal ovarian surface epithelial samples.
Mann–Whitney test.
Table 4. Molecular classification of ovarian tumours used in different sets of samples.
| Sample set | Gene classifiers | Correctly classified (n/%) | Errors | Nonclassified | Ratio (error/correct) |
|---|---|---|---|---|---|
| Training set (n=65) | 4888 genes | 33/51 | 8 | 24 | 0.24 |
| 130 genes | 52/80 | 8 | 5 | 0.15 | |
| 18 genes | 60/92 | 3 | 2 | 0.05 | |
| 130 genes | 132/96 | 4 | 1 | 0.06 | |
| Test set tissue (n=137) | 18 genes | 132/96 | 2 | 3 | 0.03 |
| Test set (n=23) | 130 genes | 18/78 | 2 | 3 | 0.11 |
| 18 genes | 20/87 | 1 | 2 | 0.05 |
k-neighbour class prediction of different sample sets. Cutoff 0.2.
Validation by Q-PCR
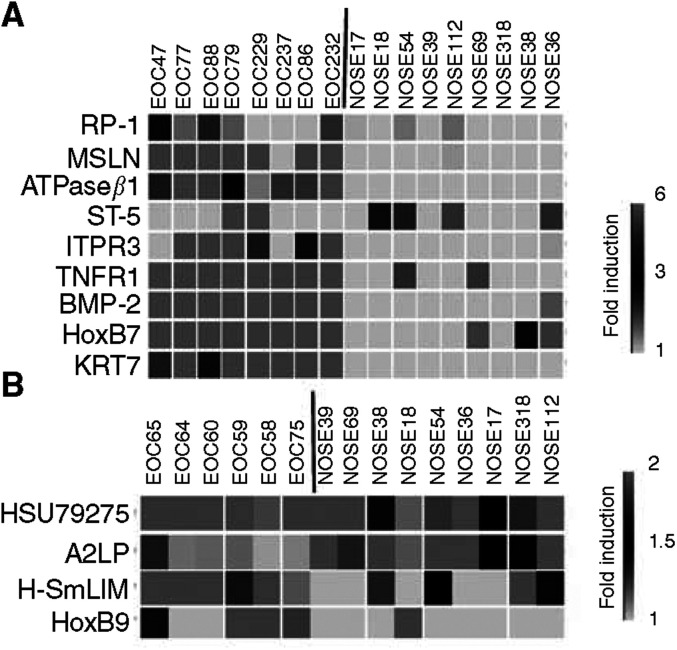
To validate the differential expression observed by DNA microarray, we tested the expression of 13 markers encoding known proteins by real-time Q-PCR. This set of 13 markers was composed of a subset of 10 markers selected from the classifier analysis (MSLN, RP-1, ITPR3, HoxB7, ATPaseβ1, ST-5, HoxB9, SmLIM, A2LP and HSU79271) in addition to three genes (TNFRI, KRT7 and BMP-2) that not only differentiate NOSEs and EOCs but also reveal differences between ascites and solid tumours (Figure 1 and Supplementary data 1). RNA from primary cultures of nine NOSE, eight malignant EOC or six LMP tumours already used for the profiling analysis was randomly chosen as templates for these assays. As shown in Figure 3, in general, Q- PCR results were consistent with expression microarray data patterns. A significant difference (P<0.05) was noticed between the expression levels of NOSE and EOC for each of the tested genes, with the exception of RP-1 and SmLIM (Figure 3A and B, respectively).
Figure 3.
Validation by real-time Q-PCR. A 2 μg portion of RNA extracted from primary culture was retro-transcribed and used for real-time Q-PCR using specific primers for RP-1, MSLN, ATPaseβ1, HoxB7, ST-5, ITPR3, TNFR1, KRT7 and BMP-2 in eight malignant samples and nine NOSE (normal ovarian surface epithelia) (A), A2RP, HSU79271, HoxB9 and SmLIM in six LMP (low malignant potential) samples and nine NOSE (B). Each expression level was normalised to that of the control RNA. Relative fold change expression is the ratio of the 61 NOSE gene expression to that of other samples. Owing to the downregulated profile of ST-5 gene expression, PCR was performed using EOC908 as reference. Green colour represents expression ratio lower than 1, black represents expression ratio equal to 1 and red represents expression ratio higher than 1.
We chose the 11 genes that were validated by Q-PCR as differentially expressed between NOSE and EOC, and tested whether their relative RNA expression monitored by Q-PCR could classify the samples using the ‘leave-one-out’ cross-validation scheme. All tumours were correctly classified; however, two NOSE samples were indistinguishable from LMP, giving a total of 87% accuracy. Even with a small set of samples, these results showed that PCR classification using a smaller set of marker genes could reach the accuracy rate obtained by microarray data.
Immunochemical expression of ATPaseβ1, TNFR1, BMP-2 and Msln in ovarian tumours
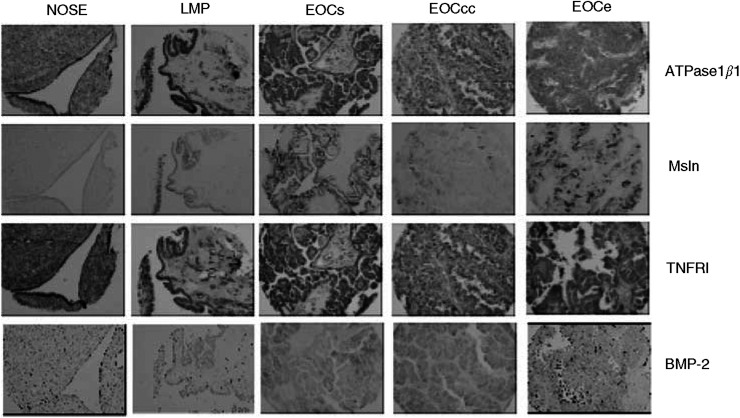
We also determined whether the protein encoded by the classifier genes had similar classifying potential. For this purpose, we used immunohistochemical assays on a tissue array containing 20 NOSE and 69 EOC cores (Table 1). This assay was restricted by the commercial availability of antibodies and thus we were not able to test any of the seven of the 10 LMP classifying genes. Among the 11 classifiers of highly malignant tumours, we were able to test ATPaseβ1 and Msln expression. We also immunostained the tissue array with keratin 19 as a positive control to visualise epithelial cells. ATPaseβ1 was expressed at a high level in both NOSE and EOC (Figure 4). Although IHC detected apparently high levels of expression of ATPase in both tissues, it was unable to discriminate the samples based on the intensity of staining. In contrast to ATPaseβ1, Msln protein was weakly expressed in some normal epithelial cells (Table 5) and more strongly in cancerous cells (Figure 4). As already described (Chang et al, 2003; Ordonez, 2003; Drapkin et al, 2004), Msln staining was observed on the apical layer of tumour tissues (Figure 4). The intensity of staining was correlated with the highest-grade tumours, where a significant difference was observed between normal epithelia and grade 2 or grade 3 tumours (P<0.03; Table 5). However, all serous tumours stained for Msln regardless of the malignancy (Table 5), supporting the idea that Msln is a marker of serous tumours independently of the grade and stage.
Figure 4.
Immunohistochemistry of NOSE and EOC tissues. Expression of ATPaseβ1, MSLN, TNFR1 and BMP-2 in normal ovarian surface epithelium (NOSE), low-malignant (LMP) and high-malignant serous (EOCs), clear cell (EOCcc) and endometrioid (EOCe) ovarian cancer tissues. Keratin 19 is strongly expressed in all three tissue types. ATPaseβ1 and TNFRI are expressed in all tissue types. Mesothelin and BMP-2 are weakly expressed in NOSE tissues, whereas they are more strongly expressed in EOC tissues. Note the apical expression of Msln.
Table 5. Immunohistochemical staining of an ovarian tissue array with anti-mesothelin and anti-BMP-2 antibodies.
| P a | 0 (n) | 1+ (n) | 2+ (n) | 3+ (n) | 4+(n) | ||
|---|---|---|---|---|---|---|---|
| Mesothelin | Normal (n=20) | 7 | 6 | 6 | 1 | 0 | |
| (n=4) | 0.01 | 0 | 0 | 1 | 1 | 2 | |
| Grade 1 (n=18) | 0.25 | 8 | 2 | 6 | 3 | 0 | |
| Grade 2 (n=17) | 0.03 | 7 | 2 | 5 | 3 | ||
| Grade 3 (n=24) | 0.01 | 7 | 3 | 2 | 10 | 2 | |
| Clear cells (n=17) | 0.41 | 10 | 0 | 3 | 4 | 0 | |
| Endometrioid (n=27) | 0.25 | 13 | 3 | 4 | 6 | 1 | |
| Serous (n=21) | <0.01 | 1 | 2 | 3 | 9 | 6 | |
| Mixed (n=5) | 0.14 | 1 | 1 | 1 | 1 | 1 | |
| Total tumours (n=70) | 0.07 | 25 | 6 | 11 | 20 | 8 | |
| BMP-2 | Normal (n=20) | 8 | 0 | 7 | 5 | 0 | |
| Grade LMP serous (n=4) | 0.25 | 1 | 1 | 0 | 0 | 2 | |
| Grade 1 (n=18) | 0.20 | 8 | 0 | 1 | 4 | 5 | |
| Grade 2 (n=17) | <0.01 | 1 | 1 | 2 | 3 | 10 | |
| Grade 3 (n=24) | <0.01 | 3 | 1 | 4 | 4 | 12 | |
| Clear cells (n=17) | <0.01 | 1 | 0 | 1 | 0 | 16 | |
| Endometrioid (n=27) | 0.04 | 8 | 1 | 3 | 5 | 10 | |
| Serous (n=21) | 0.06 | 5 | 2 | 3 | 5 | 6 | |
| Mixed (n=5) | 0.25 | 2 | 0 | 0 | 1 | 2 | |
| Total tumours (n=70) | 0.05 | 16 | 3 | 7 | 11 | 34 |
Bold values highlight the positive cancerous samples and the negative normal epithelia samples.
LMP=low malignant potential.
t-Test.
We also tested the expression of TNFR1 and BMP-2, two upregulated genes in tumour ascites cells relative to NOSE samples. As observed with ATPaseβ1, TNFR1 was expressed at a high level in both NOSE and EOC samples (Figure 4) and was unable to discriminate the samples based on the intensity of staining. In contrast, when expressed in NOSE tissues, the expression of BMP-2 was globally weaker than in tumour tissues (Table 5 and Figure 4). In contrast to Msln expression, no correlation between tumour grade and BMP-2 expression was observed, and the clear cell tumours showed a stronger staining than endometrioid and serous tissues. We also estimated the performance of BMP-2 as an individual marker of ovarian cancer. Threshold of moderate staining (3+) yielded the best possible sensitivity and specificity values to predict the tumoral status potential.
DISCUSSION
Owing to the complexity of ovarian cancer and, in particular, the presence of mixed subtypes, there is considerable interest in defining a molecular signature for ovarian cancer. Using microarray analysis, we were able to identify distinctive profiles of gene expression of ovarian cancer cells. These profiles distinguished LMP tumours, malignant tumours and malignant cells from ovarian ascites. These three groups of ovarian cancer cells are currently distinguishable by their invasive potential but not by histopathologic subtypes as previously observed (Schaner et al, 2003). Low malignant potential tumours are the least aggressive tumours of the studied groups and rarely invade the peritoneal cavity or migrate to the omentum. In contrast to solid malignant tumours that invade the stroma of the ovary and may be associated with local sites of progression, ascites tumour cells may implant at distal sites in the peritoneal cavity. The ability, in our study, to distinguish solid tumours vs ascites based on gene expression signatures may reflect different biological characteristics of these cells associated with the invasiveness and migration potential, as highlighted by the number of genes associated with angiogenesis, adhesion and metastasis processes (Table 2).
We also evaluated the classification ability of the expression of several genes detected by microarray. The predictive property of the gene set classifier may have considerable clinical importance if validated in a large set of tissue samples. Indeed, we were able to correctly classify approximately 90% of the samples. This represents a result comparable to what has been obtained with other types of cancer (Golub et al, 1999; Beer et al, 2002; Shipp et al, 2002; van’t Veer et al, 2002; Dyrskjot et al, 2003; Gordon et al, 2003; Simon, 2003), and represents the first report of this type in ovarian cancer. The success of our approach may in part be explained by the choice of the model system, which does not rely on primary undissected tumour tissue that may contain several cell types. In particular, we have previously demonstrated that even short-term passage of primary cultures results in an enriched homogeneous cell population (Lounis et al, 1994). The absence of non-malignant contaminating cells in our samples has probably allowed a strict selection of genes specifically expressed in epithelial cells. The second reason is the strategy of using the combination of small sets of specific genes to build a classifier able to differentiate simultaneously low- and high-grade malignant tumours. Gene-level expressions observed by Q-PCR confirmed the usefulness of this method and allowed for the distinction of tumour samples, which not only validated the biological relevance of the gene markers but also supported the use of Q-PCR as a new diagnostic/prognostic tool for determining tumour class. Although tumour classification based on gene expression detected by RT–PCR has already been successfully applied in mesothelia cancer (Gordon et al, 2003), the validity of this method for ovarian cancer has to be confirmed prospectively in a larger set of patient specimens.
Interestingly, our LMP tumours showed the least distinct profile in comparison to NOSE (Figure 1), which correlates with the weak aggressive potential of these tumours and the favourable prognostic for the patient. The present results correlate well with recent attempts to distinguish LMP and solid malignant tumours using a microarray and gene profiling approach (Gilks et al, 2005; Meinhold-Heerlein et al, 2005; Ouellet et al, 2005). Here, we detected only a very small set of genes differentially expressed in LMP tumours. Those genes are genes encoding for unknown proteins such as HSU79275, U60269, HG830-HT830, and genes encoding for proteins without a clear function related to oncogenesis, with the exception of the gene SAS (sarcoma amplified sequence). Owing to its close localisation with MDM2, an inhibitor of p53, the SAS oncogene is often associated with the amplification of MDM2 in human sarcomas (Meltzer et al, 1991). In ovarian cancer expression, MDM2 is more characteristic of serous LMP (Palazzo et al, 2000). A deeper analysis would be necessary to determine the potential involvement of SAS and MDM2 and their prognostic value in serous LMP cancer. In addition, the small number of genes detected as differentially expressed between LMP and NOSE suggests that these tumours have a very similar profile to NOSE cells, and also suggests that a better molecular distinction between NOSE and LMP will need further investigation.
In the context of tumour suppressors involved in ovarian cancer, much attention has focused on the role of BRCA and p53. Whereas p53 mutation appears to be frequent (Marks et al, 1991; Okamoto et al, 1991), BRCA mutation occurs in less than 10% in all diagnosed cases (Ford et al, 1994; Cass et al, 2003) and represents a small minority of ovarian cancers. Here, we identified ST-5/HTS1 as a tumour suppressor gene downregulated in the majority of ovarian primary cultures derived from malignant tumours. ST-5 has been initially identified as a HeLa tumour suppression gene (Lichy et al, 1992; Amid et al, 2001). Interestingly, HeLa cells are derived from an uterine tumour, which suggests that ST-5 may be hormonally regulated. However, little is known about the regulation and the expression of this tumour suppressor. Future interests will allow a better understanding of the role of ST-5 in the gynaecological female tract cancers such as ovarian and uterine cancers.
Among the genes differentially expressed in malignant EOC is Msln. Mesothelin mRNA has previously been described as overexpressed in ovarian and mesothelioma cancer tissues, and the gene product is referred to a marker of these two cancers (Ordonez, 2003; Drapkin et al, 2004; McIntosh et al, 2004). However, these studies analysed the Msln protein expression using a small number of samples, and usually limited their analysis to serous ovarian cancer samples. We extended these observations on a larger set of samples also containing endometrioid and clear cell tissues with different tumour grades. In contrast to the general statement that Msln is a specific ovarian cancer marker, here we observed that normal epithelial cells can at times express Msln and a weak or absent expression is observed in both low-grade endometrioid and clear cell tumours. Our extended analysis would thus emphasise that no unique protein marker may be appropriate to classify such a heterogeneous disease, and reinforces the notion that a combination of protein markers is possibly necessary to allow the correct distinction of normal and cancerous ovarian epithelial cells. Our results support this idea but do not rule out the idea that Msln may be an appropriate ovarian cancer marker in association with other markers, such as BMP-2, that complement staining in low-grade endometrioid and clear cell tumours.
In conclusion, our results indicate that a molecular classification system, based on the statistical analysis of gene expression profiling, is a useful approach for tumour subgrouping and the discovery of new molecular markers. These observations articulate a new area of research in the understanding of ovarian cancer as well as illuminating new therapeutic strategies. Combination of oligo-microarray, RT–PCR and tissue array linked to clinical and pathology data will facilitate rapid characterisation of candidate markers.
Acknowledgments
We are grateful to the staff and patients at the Department of Onco-Gynecology at the Hôpital Notre-Dame de Montréal for providing the samples. We thank Lise Portelance, Manon de Ladurantaye, Louise Champoux, Stéphanie Girard, and Dr Herve Koumakpahi for their assistance. We are grateful to Dr Mario Fillion for RNA amplification and Dr Ali Filali, for bioinformation. We also thank Dr Nadia Benhachenhou and the ‘oncotree group’ for useful discussions. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to A-MM-M, PNT, DMP and TJH. Tumor banking was supported by the Banque de tissus et de données of the Réseau de recherche sur le cancer of the Fonds de la recherche en santé du Québec (FRSQ). VO was supported by studentships from the CIHR and Canderel fund of the Institut du cancer de Montréal. FR was supported by a Wyeth-CIHR-sponsored fellowship and PNT is a recipient of a Frazer, Monat and Mc Pherson Scholarship and the Stewart Fellowship in Research/Clinical Hematology and Oncology. TJH is recipient of an Investigator Award from CIHR and a Clinician-scientist Award in Translational Research from the Burroughs Wellcome Fund. DMP is a recipient of a Chercheur-Clinicien Senior and A-MM-M is a recipient of a Chercheur National, all fellowships provided by the FRSQ.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Adib TR, Henderson S, Perrett C, Hewitt D, Bourmpoulia D, Ledermann J, Boshoff C (2004) Predicting biomarkers for ovarian cancer using gene-expression microarrays. Br J Cancer 90: 686–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amid C, Bahr A, Mujica A, Sampson N, Bikar SE, Winterpacht A, Zabel B, Hankeln T, Schmidt ER (2001) Comparative genomic sequencing reveals a strikingly similar architecture of a conserved syntenic region on human chromosome 11p15.3 (including gene ST5) and mouse chromosome 7. Cytogenet Cell Genet 93: 284–290 [DOI] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8: 816–824 [DOI] [PubMed] [Google Scholar]
- Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY (2003) Improved survival in women with BRCA-associated ovarian carcinoma. Cancer 97: 2187–2195 [DOI] [PubMed] [Google Scholar]
- Chang XH, Cheng YX, Zhang XY, Cui H, Feng J (2003) [Monitoring novel ovarian carcinoma associated genes using cDNA expression microarray]. Zhonghua Fu Chan Ke Za Zhi 38: 322–324 [PubMed] [Google Scholar]
- Donninger H, Bonome T, Radonovich M, Pise-Masison CA, Brady J, Shih JH, Barrett JC, Birrer MJ (2004) Whole genome expression profiling of advance stage papillary serous ovarian cancer reveals activated pathways. Oncogene 23: 8065–8077 [DOI] [PubMed] [Google Scholar]
- Drapkin R, Crum CP, Hecht JL (2004) Expression of candidate tumor markers in ovarian carcinoma and benign ovary: evidence for a link between epithelial phenotype and neoplasia. Hum Pathol 35: 1014–1021 [DOI] [PubMed] [Google Scholar]
- Dyrskjot L, Thykjaer T, Kruhoffer M, Jensen JL, Marcussen N, Hamilton-Dutoit S, Wolf H, Orntoft TF (2003) Identifying distinct classes of bladder carcinoma using microarrays. Nat Genet 33: 90–96 [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Bishop DT, Narod SA, Goldgar DE (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343: 692–695 [DOI] [PubMed] [Google Scholar]
- Gilks CB, Vanderhyden BC, Zhu S, van de Rijn M, Longacre TA (2005) Distinction between serous tumors of low malignant potential and serous carcinomas based on global mRNA expression profiling. Gynecol Oncol 96: 684–694 [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286: 531–537 [DOI] [PubMed] [Google Scholar]
- Gordon GJ, Richards WG, Sugarbaker DJ, Jaklitsch MT, Bueno R (2003) A prognostic test for adenocarcinoma of the lung from gene expression profiling data. Cancer Epidemiol Biomarkers Prev 12: 905–910 [PubMed] [Google Scholar]
- Hibbs K, Skubitz KM, Pambuccian SE, Casey RC, Burleson KM, Oegema TR, Jr, Thiele JJ, Grindle SM, Bliss RL, Skubitz AP (2004) Differential gene expression in ovarian carcinoma: identification of potential biomarkers. Am J Pathol 165: 397–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruk PA, Maines-Bandiera SL, Auersperg N (1990) A simplified method to culture human ovarian surface epithelium. Lab Invest 63: 132–136 [PubMed] [Google Scholar]
- Lancaster JM, Dressman HK, Whitaker RS, Havrilesky L, Gray J, Marks JR, Nevins JR, Berchuck A (2004) Gene expression patterns that characterize advanced stage serous ovarian cancers. J Soc Gynecol Investig 11: 51–59 [DOI] [PubMed] [Google Scholar]
- Le Page C, Provencher D, Maugard CM, Ouellet V, Mes-Masson AM (2004) Signature of a silent killer: expression profiling in epithelial ovarian cancer. Expert Rev Mol Diagn 4: 157–167 [DOI] [PubMed] [Google Scholar]
- Lee BC, Cha K, Avraham S, Avraham HK (2004) Microarray analysis of differentially expressed genes associated with human ovarian cancer. Int J Oncol 24: 847–851 [PubMed] [Google Scholar]
- Lichy JH, Modi WS, Seuanez HN, Howley PM (1992) Identification of a human chromosome 11 gene which is differentially regulated in tumorigenic and nontumorigenic somatic cell hybrids of HeLa cells. Cell Growth Differ 3: 541–548 [PubMed] [Google Scholar]
- Lounis H, Provencher D, Godbout C, Fink D, Milot MJ, Mes-Masson AM (1994) Primary cultures of normal and tumoral human ovarian epithelium: a powerful tool for basic molecular studies. Exp Cell Res 215: 303–309 [DOI] [PubMed] [Google Scholar]
- Lu KH, Patterson AP, Wang L, Marquez RT, Atkinson EN, Baggerly KA, Ramoth LR, Rosen DG, Liu J, Hellstrom I, Smith D, Hartmann L, Fishman D, Berchuck A, Schmandt R, Whitaker R, Gershenson DM, Mills GB, Bast Jr RC (2004) Selection of potential markers for epithelial ovarian cancer with gene expression arrays and recursive descent partition analysis. Clin Cancer Res 10: 3291–3300 [DOI] [PubMed] [Google Scholar]
- Marks JR, Davidoff AM, Kerns BJ, Humphrey PA, Pence JC, Dodge RK, Clarke-Pearson DL, Iglehart JD, Bast Jr RC, Berchuck A (1991) Overexpression and mutation of p53 in epithelial ovarian cancer. Cancer Res 51: 2979–2984 [PubMed] [Google Scholar]
- McIntosh MW, Drescher C, Karlan B, Scholler N, Urban N, Hellstrom KE, Hellstrom I (2004) Combining CA 125 and SMR serum markers for diagnosis and early detection of ovarian carcinoma. Gynecol Oncol 95: 9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhold-Heerlein I, Bauerschlag D, Hilpert F, Dimitrov P, Sapinoso LM, Orlowska-Volk M, Bauknecht T, Park TW, Jonat W, Jacobsen A, Sehouli J, Luttges J, Krajewski M, Krajewski S, Reed JC, Arnold N, Hampton GM (2005) Molecular and prognostic distinction between serous ovarian carcinomas of varying grade and malignant potential. Oncogene 24: 1053–1065 [DOI] [PubMed] [Google Scholar]
- Meltzer PS, Jankowski SA, Dal Cin P, Sandberg AA, Paz IB, Coccia MA (1991) Identification and cloning of a novel amplified DNA sequence in human malignant fibrous histiocytoma derived from a region of chromosome 12 frequently rearranged in soft tissue tumors. Cell Growth Differ 2: 495–501 [PubMed] [Google Scholar]
- Neibergs HL, Hein DW, Spratt JS (2002) Genetic profiling of colon cancer. J Surg Oncol 80: 204–213 [DOI] [PubMed] [Google Scholar]
- Okamoto A, Sameshima Y, Yokoyama S, Terashima Y, Sugimura T, Terada M, Yokota J (1991) Frequent allelic losses and mutations of the p53 gene in human ovarian cancer. Cancer Res 51: 5171–5176 [PubMed] [Google Scholar]
- Ordonez NG (2003) Application of mesothelin immunostaining in tumor diagnosis. Am J Surg Pathol 27: 1418–1428 [DOI] [PubMed] [Google Scholar]
- Ouellet V, Provencher DM, Maugard CM, Le Page C, Ren F, Lussier C, Novak J, Ge B, Hudson TJ, Tonin PN, Mes-Masson AM (2005) Discrimination between serous low malignant potential and invasive epithelial ovarian tumors using molecular profiling. Oncogene 24: 4672–4687 [DOI] [PubMed] [Google Scholar]
- Palazzo JP, Monzon F, Burke M, Hyslop T, Dunton C, Barusevicius A, Capuzzi D, Kovatich AJ (2000) Overexpression of p21WAF1/CIP1 and MDM2 characterizes serous borderline ovarian tumors. Hum Pathol 31: 698–704 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR (2001) Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA 98: 15149–15154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, Iyer V, Jeffrey SS, Van de Rijn M, Waltham M, Pergamenschikov A, Lee JC, Lashkari D, Shalon D, Myers TG, Weinstein JN, Botstein D, Brown PO (2000) Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 24: 227–235 [DOI] [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O’Brien TJ, Tian E, Cannon MJ, Shaughnessy Jr J, Pecorelli S (2004) Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer 112: 14–25 [DOI] [PubMed] [Google Scholar]
- Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, Skomedal H, Tu IP, Hernandez-Boussard T, Johnson SW, O’Dwyer PJ, Fero MJ, Kristensen GB, Borresen-Dale AL, Hastie T, Tibshirani R, van de Rijn M, Teng NN, Longacre TA, Botstein D, Brown PO, Sikic BI (2003) Gene expression patterns in ovarian carcinomas. Mol Biol Cell 14: 4376–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, Reed H, Ellenson LH, Giordano TJ, Fearon ER, Hanash SM, Cho KR (2002) Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res 62: 4722–4729 [PubMed] [Google Scholar]
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR (2002) Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med 8: 68–74 [DOI] [PubMed] [Google Scholar]
- Simon R (2003) Using DNA microarrays for diagnostic and prognostic prediction. Expert Rev Mol Diagn 3: 587–595 [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 96: 2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536 [DOI] [PubMed] [Google Scholar]
- Warrenfeltz S, Pavlik S, Datta S, Kraemer ET, Benigno B, McDonald JF (2004) Gene expression profiling of epithelial ovarian tumours correlated with malignant potential. Mol Cancer 3: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM (2001) Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA 98: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng J, Cheng Y, Yao Y, Ye X, Fu T, Cheng H (2005) Characterization of differentially expressed genes in ovarian cancer by cDNA microarrays. Int J Gynecol Cancer 15: 50–57 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.