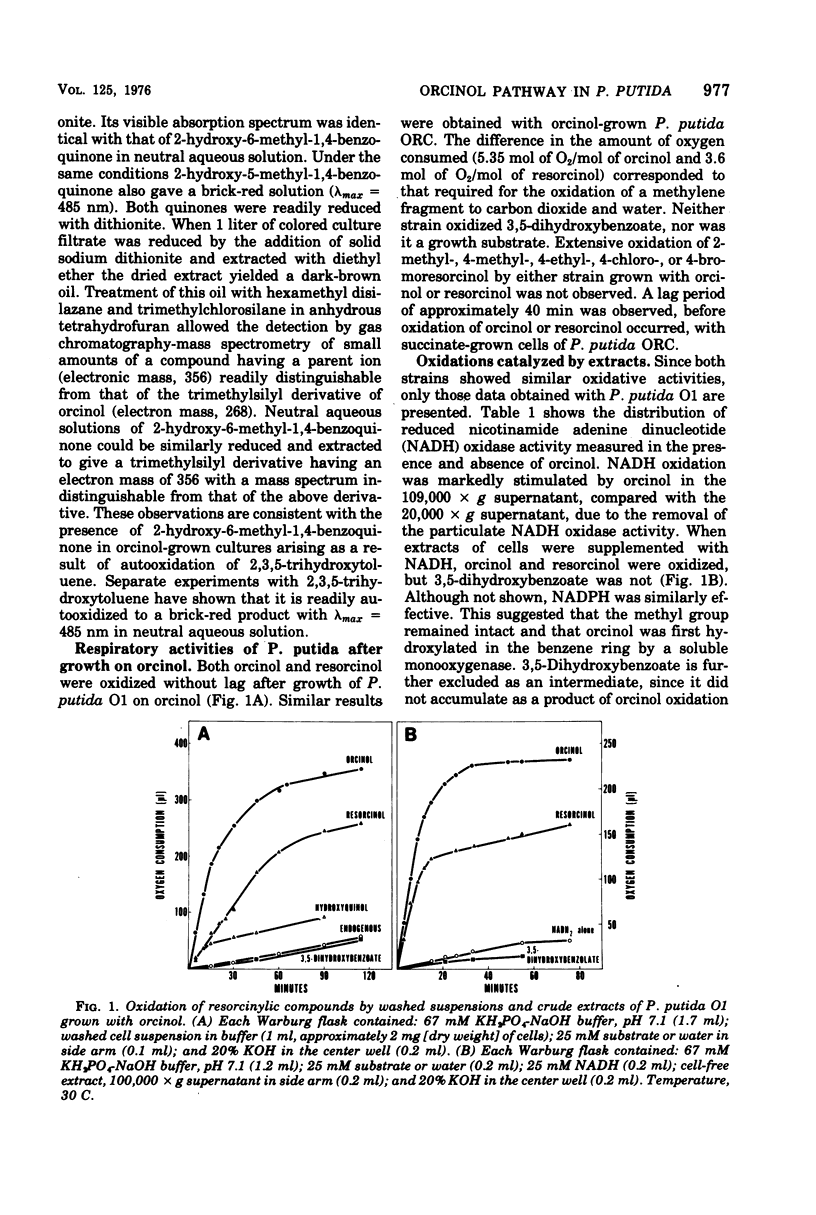
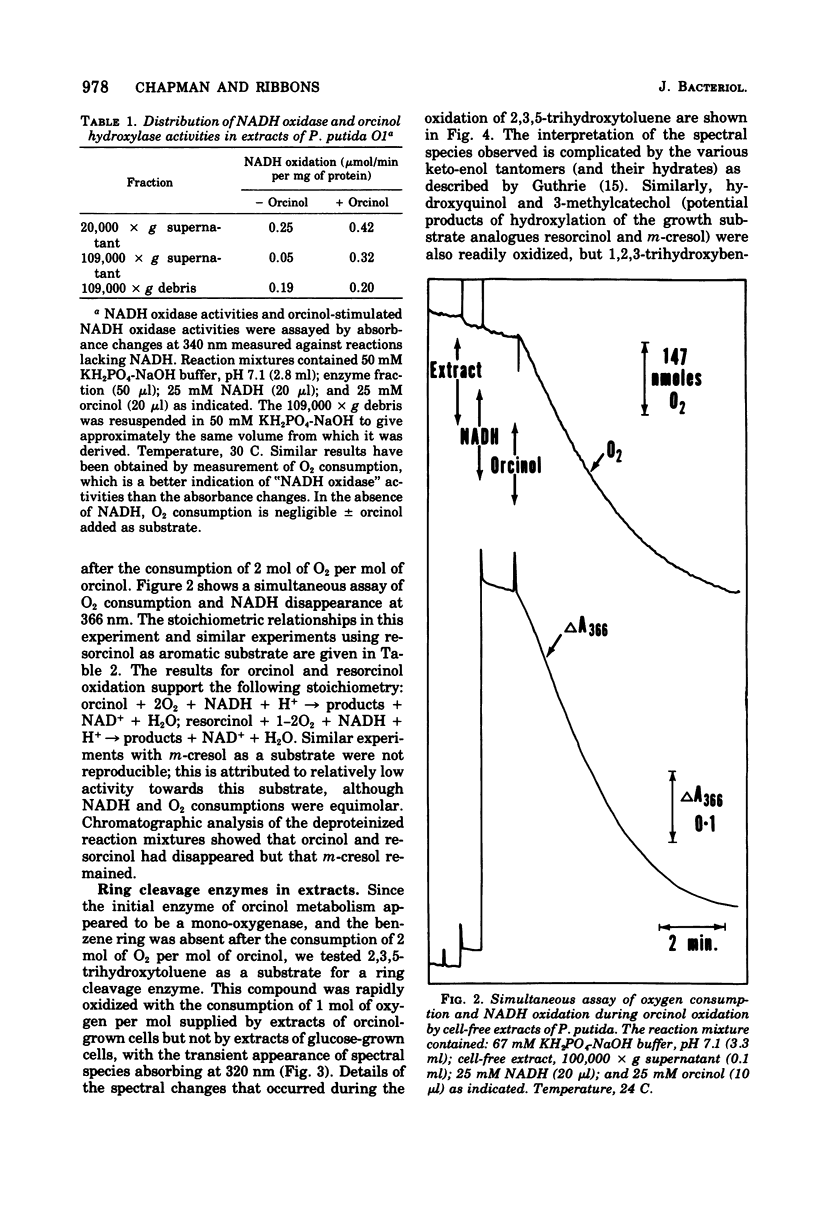
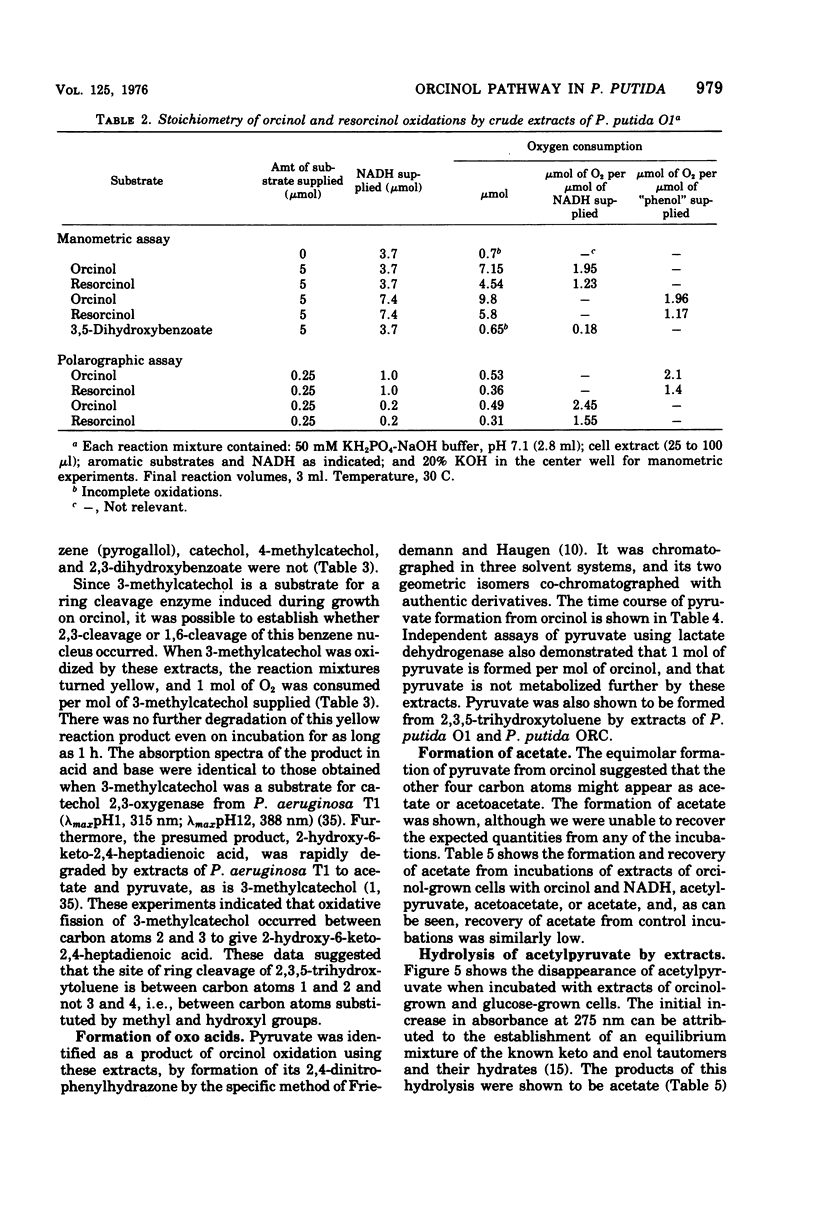
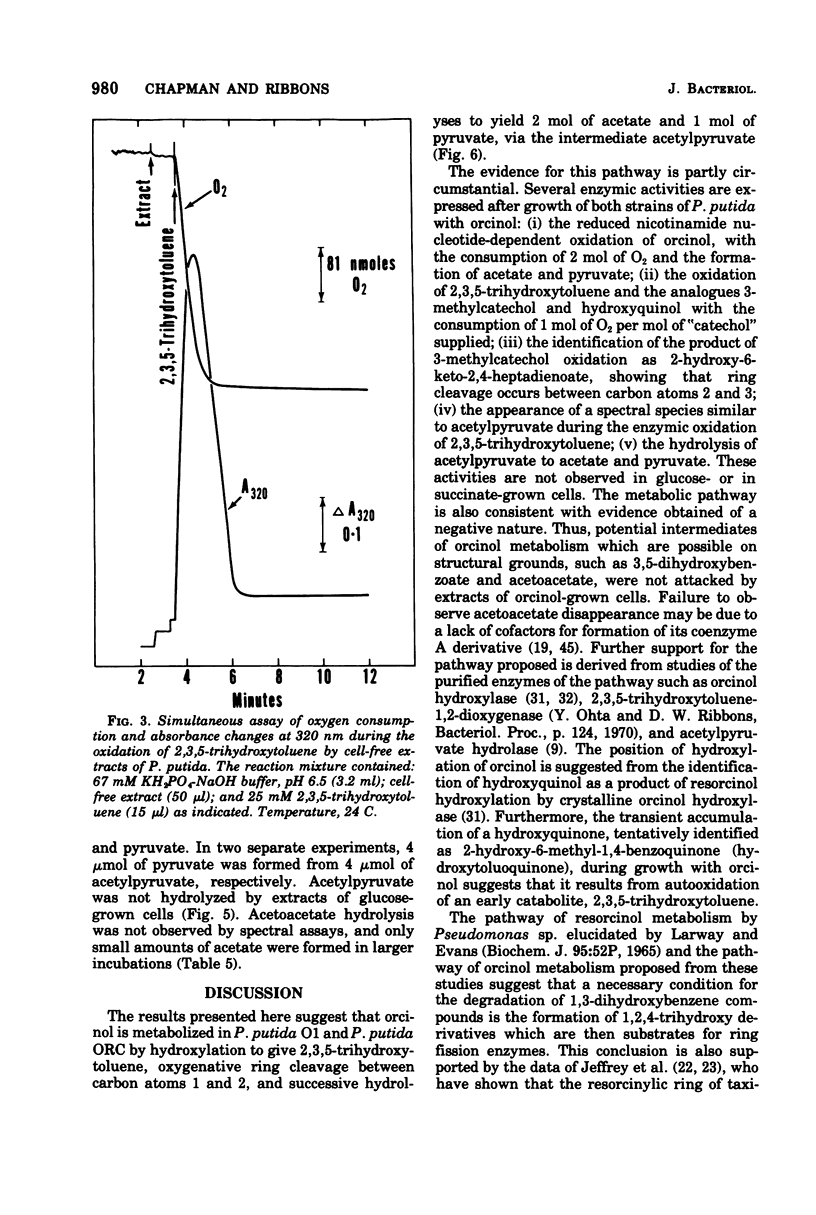
Abstract
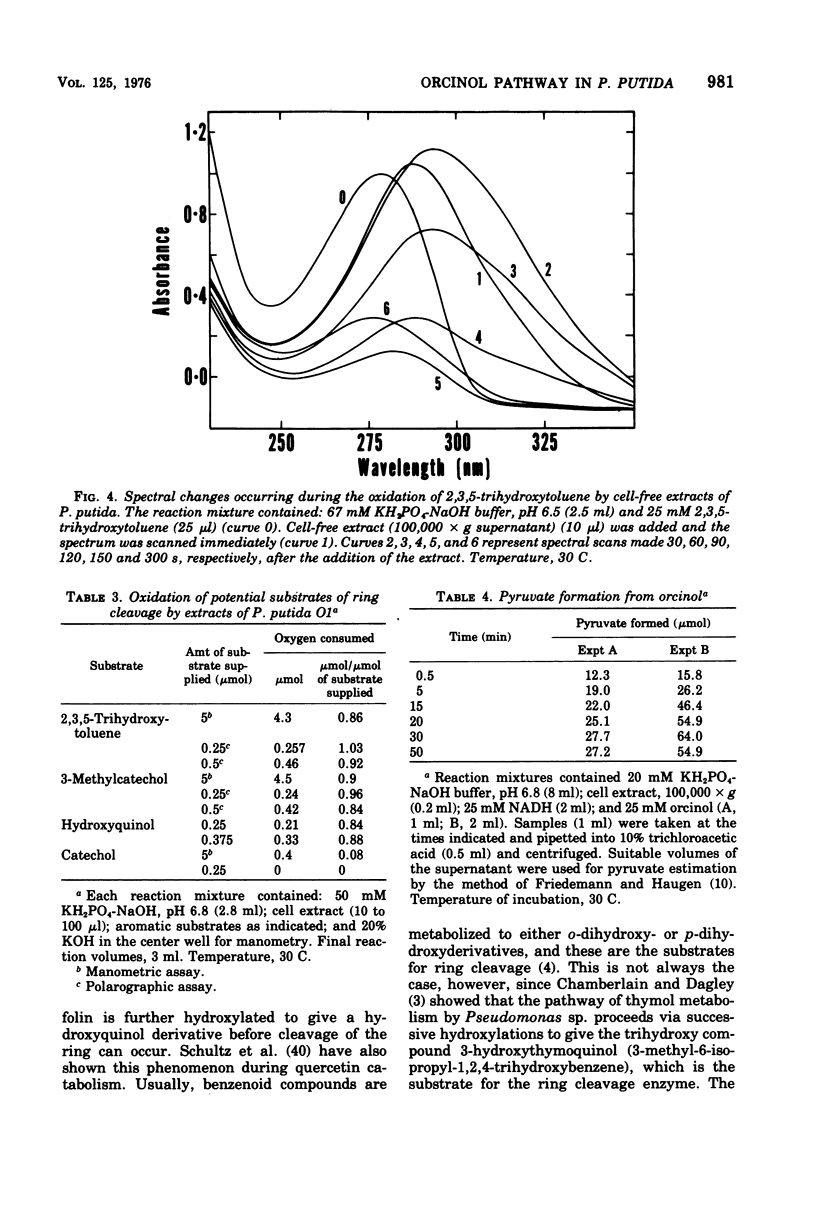
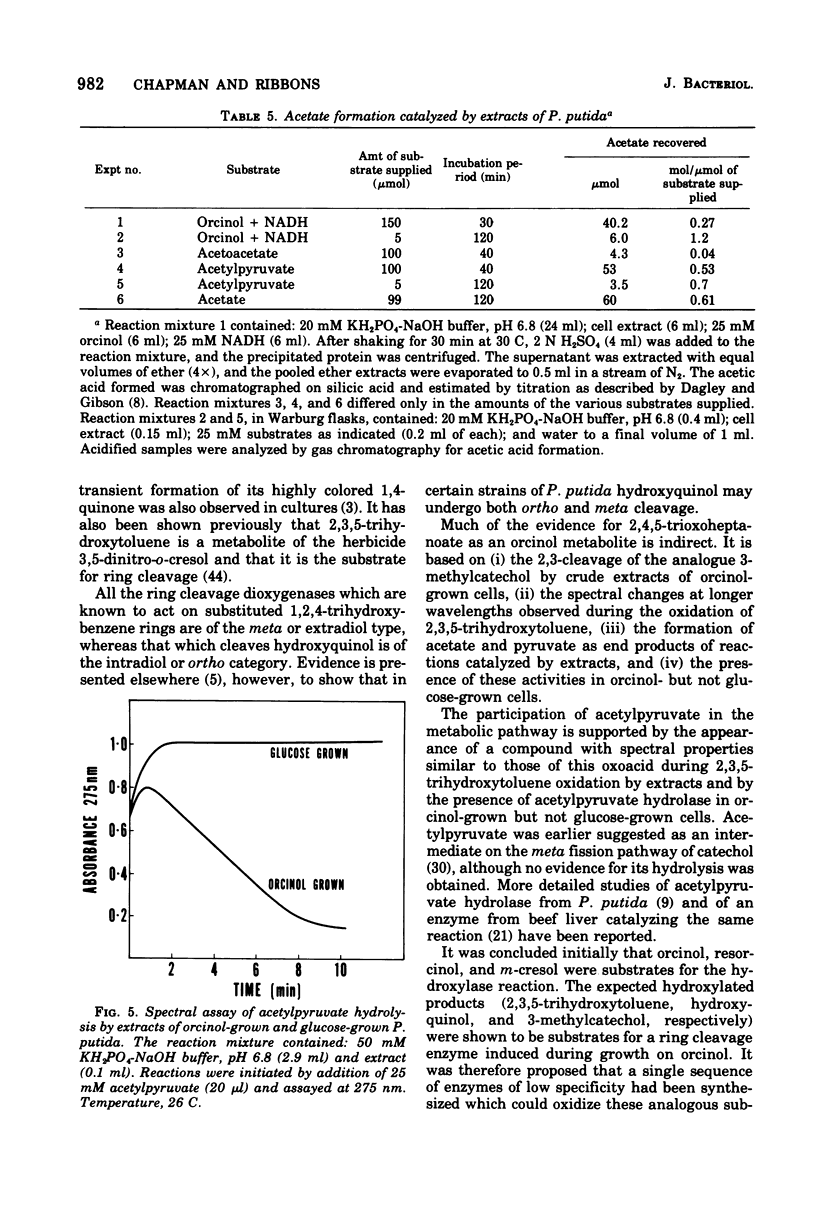
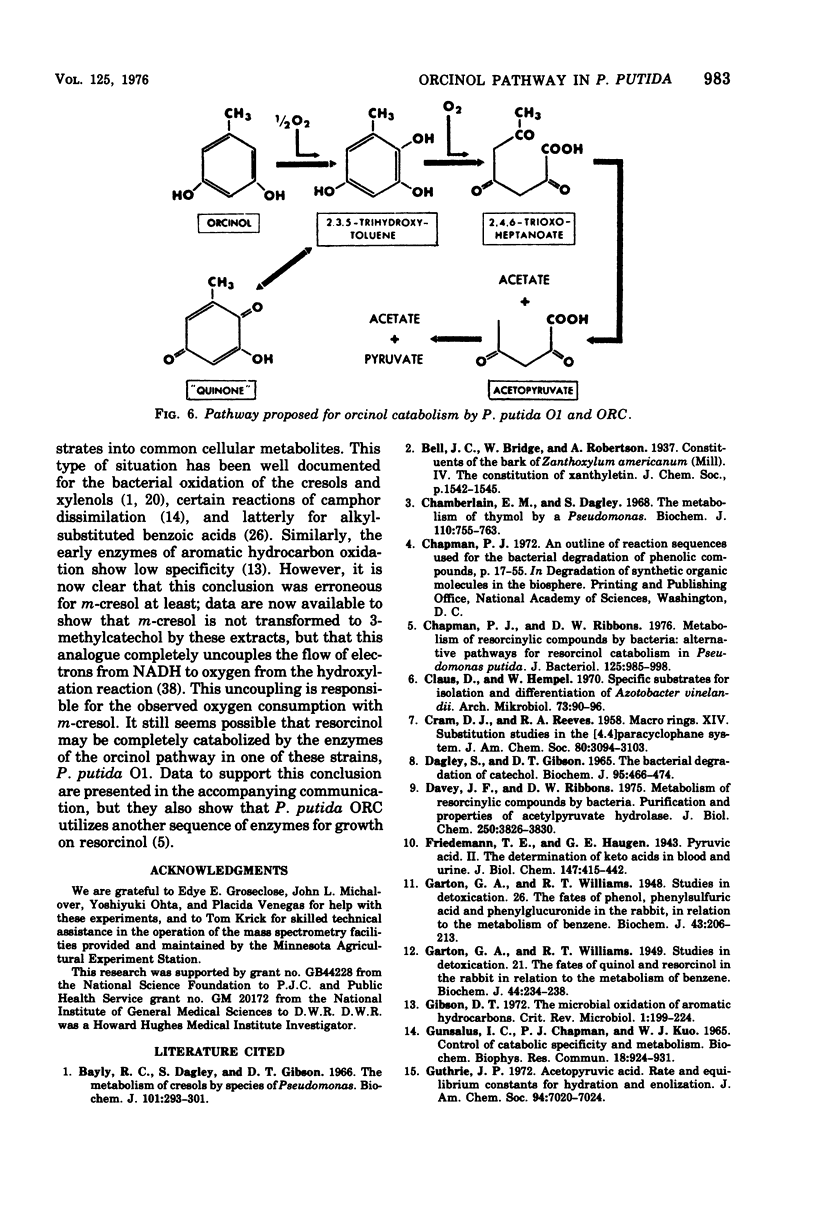
Enrichment cultures yielded two strains of Pseudomonas putida capable of growth with orcinol (3,5-dihydroxytoluene) as the sole source of carbon. Experiments with cell suspensions and cell extracts indicate that orcinol is metabolized by hydroxylation of the benzene ring followed successively by ring cleavage and hydrolyses to give 2 mol of acetate and 1 mol of pyruvate per mol of orcinol as shown: orcinol leads to 2,3,5-trihydroxytoluene leads to 2,4,6-trioxoheptanoate leads to acetate + acetylpyruvate leads to acetate + pyruvate. Evidence for this pathway is based on: (i) high respiratory activities of orcinol-grown cells towards 2,3,5-trihydroxytoluene; (ii) transient accumulation of a quinone, probably 2-hydroxy-6-methyl-1,4-benzoquinone, during grouth with orcinol; (iii) formation of pyruvate and acetate from orcinol, 2,3,5-trihydroxytoluene, and acetylpyruvate catalyzed by extracts of orcinol, but not by succinate-grown cells; (iv) characterization of the product of oxidation of 3-methylcatechol (an analogue of 2,3,5-trihydroxytoluene) showing that oxygenative cleavage occurs between carbons bearing methyl and hydroxyl substituents; (v) transient appearance of a compound having spectral properties similar to those of acetylpyruvate during 2,3,5-trihydroxytoluene oxidation by extracts of orcinol-grown cells. Orcinol hydroxylase exhibits catalytic activity when resorcinol or m-cresol is substituted for orcinol; hydroxyquinol and 3-methylcatechol are substrates for the ring cleavage enzyme 2,3,5-trihydroxytoluene-1,2-oxygenase. The enzymes of this pathway are induced by growth with orcinol but not with glucose or succinate.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayly R. C., Dagley S., Gibson D. T. The metabolism of cresols by species of Pseudomonas. Biochem J. 1966 Nov;101(2):293–301. doi: 10.1042/bj1010293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain E. M., Dagley S. The metabolism of thymol by a Pseudomonas. Biochem J. 1968 Dec;110(4):755–763. doi: 10.1042/bj1100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman P. J., Ribbons D. W. Metabolism of resorcinylic compounds by bacteria: alternative pathways for resorcinol catabolism in Pseudomonas putida. J Bacteriol. 1976 Mar;125(3):985–998. doi: 10.1128/jb.125.3.985-998.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus D., Hempel W. Specific substrates for isolation and differentiation of Azotobacter vinelandii. Arch Mikrobiol. 1970;73(1):90–96. doi: 10.1007/BF00409955. [DOI] [PubMed] [Google Scholar]
- DAGLEY S., GIBSON D. T. THE BACTERIAL DEGRADATION OF CATECHOL. Biochem J. 1965 May;95:466–474. doi: 10.1042/bj0950466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. F., Ribbons D. W. Metabolism of resorcinylic compounds by bacteria. Purification and properties of acetylpyruvate hydrolase from Pseudomonas putida 01. J Biol Chem. 1975 May 25;250(10):3826–3830. [PubMed] [Google Scholar]
- Garton G. A., Williams R. T. Studies in detoxication. 17. The fate of catechol in the rabbit and the characterization of catechol monoglucuronide. Biochem J. 1948;43(2):206–211. [PMC free article] [PubMed] [Google Scholar]
- Garton G. A., Williams R. T. Studies in detoxication. 21. The fates of quinol and resorcinol in the rabbit in relation to the metabolism of benzene. Biochem J. 1949;44(2):234–238. [PMC free article] [PubMed] [Google Scholar]
- HIMES R. H., LYNEN F. METABOLISM OF ACETOACETATE BY ACHROMOBACTER IV S. Biochem Z. 1964 Jul 29;340:206–212. [PubMed] [Google Scholar]
- Hopper D. J., Chapman P. J. Gentisic acid and its 3- and 4-methyl-substituted homologoues as intermediates in the bacterial degradation of m-cresol, 3,5-xylenol and 2,5-xylenol. Biochem J. 1971 Mar;122(1):19–28. doi: 10.1042/bj1220019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiang H. H., Sim S. S., Mahuran D. J., Schmidt D. E., Jr Purification and properties of a diketo acid hydrolase from beef liver. Biochemistry. 1972 May 23;11(11):2098–2102. doi: 10.1021/bi00761a016. [DOI] [PubMed] [Google Scholar]
- Jeffrey A. M., Jerina D. M., Self R., Evans W. C. The bacterial degradation of flavonoids. Oxidative fission of the A-ring of dihydrogossypetin by a Pseudomonas sp. Biochem J. 1972 Nov;130(2):383–390. doi: 10.1042/bj1300383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey A. M., Knight M., Evans W. C. The bacterial degradation of flavonoids. Hydroxylation of the A-ring of taxifolin by a soil pseudomonad. Biochem J. 1972 Nov;130(2):373–381. doi: 10.1042/bj1300373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Murray K., Duggleby C. J., Sala-Trepat J. M., Williams P. A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972 Jul 24;28(3):301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- NEUFELD H. A., LATTERELL F. M., GREEN L. F., WEINTRAUB R. L. Oxidation of meta-polyhydroxyphenols by enzymes from Piricularia oryzae and Polyporus versicolor. Arch Biochem Biophys. 1958 Aug;76(2):317–327. doi: 10.1016/0003-9861(58)90157-7. [DOI] [PubMed] [Google Scholar]
- Neujahr H. Y., Varga J. M. Degradation of phenols by intact cells and cell-free preparations of Trichosporon cutaneum. Eur J Biochem. 1970 Mar 1;13(1):37–44. doi: 10.1111/j.1432-1033.1970.tb00896.x. [DOI] [PubMed] [Google Scholar]
- Ohta Y., Higgins I., Ribbons D. W. Metabolism of resorcinylic compounds by bacteria. Purification and properties of orcinol hydroxylase from Pseudomonas putida 01. J Biol Chem. 1975 May 25;250(10):3814–3825. [PubMed] [Google Scholar]
- Otha Y., Ribbons D. W. Crystallization of orcinol hydroxylase from Pseudomonas putida. FEBS Lett. 1970 Dec;11(3):189–192. doi: 10.1016/0014-5793(70)80525-7. [DOI] [PubMed] [Google Scholar]
- Ribbons D. W. Metabolism of omicron-cresol by Pseudomonas aeruginosa strain T1. J Gen Microbiol. 1966 Aug;44(2):221–231. doi: 10.1099/00221287-44-2-221. [DOI] [PubMed] [Google Scholar]
- Ribbons D. W., Ohta Y. Uncoupling of electron transport from oxygenation in the mono-oxygenase, orcinol hydroxylase. FEBS Lett. 1970 Dec 28;12(2):105–108. doi: 10.1016/0014-5793(70)80574-9. [DOI] [PubMed] [Google Scholar]
- STEFANYE D. Oxidation of metadihydroxyphenols by enzymes of Piricularia oryzae. J Bacteriol. 1961 Oct;82:617–618. doi: 10.1128/jb.82.4.617-618.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E., Engle F. E., Wood J. M. New oxygenases in the degradation of flavones and flavanones by Pseudomonas putida. Biochemistry. 1974 Apr 9;13(8):1768–1776. doi: 10.1021/bi00705a033. [DOI] [PubMed] [Google Scholar]
- Sparnins V. L., Chapman P. J., Dagley S. Bacterial degradation of 4-hydroxyphenylacetic acid and homoprotocatechuic acid. J Bacteriol. 1974 Oct;120(1):159–167. doi: 10.1128/jb.120.1.159-167.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Palleroni N. J., Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966 May;43(2):159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- Winnacker E. L., Barker H. A. Aerobic metabolism of -amino-n-butyric acid by Pseudomonas putida. Biochim Biophys Acta. 1971 May 18;237(2):284–292. doi: 10.1016/0304-4165(71)90319-9. [DOI] [PubMed] [Google Scholar]


