Abstract
Uterine serous papillary cancer (USPC) represents a rare but highly aggressive variant of endometrial cancer, the most common gynecologic tumour in women. We used oligonucleotide microarrays that interrogate the expression of some 10 000 known genes to profile 10 highly purified primary USPC cultures and five normal endometrial cells (NEC). We report that unsupervised analysis of mRNA fingerprints readily distinguished USPC from normal endometrial epithelial cells and identified 139 and 390 genes that exhibited >5-fold upregulation and downregulation, respectively, in primary USPC when compared to NEC. Many of the genes upregulated in USPC were found to represent adhesion molecules, secreted proteins and oncogenes, such as L1 cell adhesion molecule, claudin-3 and claudin-4, kallikrein 6 (protease M) and kallikrein 10 (NES1), interleukin-6 and c-erbB2. Downregulated genes in USPC included SEMACAP3, ras homolog gene family, member I (ARHI), and differentially downregulated in ovarian carcinoma gene 1. Quantitative RT–PCR was used to validate differences in gene expression between USPC and NEC for several of these genes. Owing to its potential as a novel therapeutic marker, expression of the high-affinity epithelial receptor for Clostridium perfringens enterotoxin (CPE) claudin-4 was further validated through immunohistochemical analysis of formalin-fixed paraffin-embedded specimens from which the primary USPC cultures were obtained, as well as an independent set of archival USPC specimens. Finally, the sensitivity of primary USPC to the administration of scalar doses of CPE in vitro was also demonstrated. Our results highlight the novel molecular features of USPC and provide a foundation for the development of new type-specific therapies against this highly aggressive variant of endometrial cancer.
Keywords: serous papillary uterine cancer, gene expression profiling
Uterine cancer is the most prevalent gynecologic tumour in women, with an estimated 40,100 cases and 6800 deaths in the United States in 2003 (Jemal et al, 2003). On the basis of clinical and histopathologic variables, two subtypes of endometrial carcinoma, namely Type I and Type II tumours, have been described (Bohkman, 1983). Type I endometrial cancers, which account for the majority (i.e., about 80%) of cases, are usually well differentiated and endometrioid in histology. These neoplasms are frequently diagnosed in younger women, are associated with a history of hyperestrogenism as the main risk factor, and typically have a favourable prognosis with appropriate therapy. Type II endometrial cancers are poorly differentiated tumours, often with serous papillary or clear cell histology, and are not associated with hyperestrogenic factors. Although Type II tumours account for only a minority of endometrial carcinoma, about 50% of all relapses occur in this group of patients.
High-throughput technologies for assaying gene expression, such as high-density oligonucleotide and cDNA microarrays, have recently been used in an attempt to define the genetic fingerprints of a variety of human tumours including endometrial cancers (Smid-Koopman et al, 2000; Matsushima-Nishiu et al, 2001; Moreno-Bueno et al, 2003; Risinger et al, 2003). These techniques may allow precise and accurate grouping of human tumours and have the potential to identify patients who are unlikely to be cured by conventional therapy (Sorlie et al, 2001; Rosenwald et al, 2002; Moreno-Bueno et al, 2003; Risinger et al, 2003). Consistent with this view, clinically relevant genes highly differentially expressed between Type I and Type II endometrial tumours have recently been identified (Moreno-Bueno et al, 2003; Risinger et al, 2003). Most of the differentially expressed genes in Type I tumours included genes known to be hormonally regulated during the menstrual cycle and known to be important in endometrial homeostasis (i.e., MGB2, LTF, END1, and MMP11) (Moreno-Bueno et al, 2003; Risinger et al, 2003). In contrast, Type II tumours have been shown to overexpress genes involved in the regulation of the mitotic spindle checkpoint and associated with aneuploidy and an aggressive phenotype, such as STK15, BUB1, and CCNB2 (Moreno-Bueno et al, 2003; Risinger et al, 2003).
Uterine serous papillary tumors (USPC) represent the most aggressive variant of Type II endometrial cancer and may constitute up to 10% of endometrial tumours. The microscopic criteria for diagnosis of USPC were first outlined by Hendrickson et al (1982). Pleomorphism, grade III nuclear atypia with prominent nucleoli and vesicular chromatin pattern, as well as a high mitotic activity are commonly detected in this tumour. Clinically, USPC has a propensity for early intra-abdominal and lymphatic spread even at presentation and is characterised by a highly aggressive biologic behaviour (Hendrickson et al, 1982; Nicklin and Copeland, 1996). Unlike the histologically indistinguishable high-grade serous ovarian carcinomas, USPC is a chemoresistant disease from onset, with responses to combined cisplatinum-based chemotherapy in the order of 20% and of short duration (Nicklin and Copeland, 1996). The overall 5-year survival is about 30% for all stages and the recurrence rate after surgery is extremely high (50–80%). A deeper understanding of the molecular basis of the aggressive biologic behaviour of USPC as well as the development of novel, more specific and more effective treatment modalities against this variant of endometrial cancer remain a high priority.
In this study, with the goal of identifying genes with a differential pattern of expression between USPC and normal endometrial cells (NEC) and to use this knowledge for the development of novel diagnostic and therapeutic markers against this disease, we used oligonucleotide microarrays which interrogate the expression of some 10 000 known genes to analyse gene expression profiling of 10 highly purified primary USPC cultures and five primary NEC. We report a number of genes which may readily distinguish USPC from normal endometrial epithelial cells. More importantly, these results highlight the novel molecular features of USPC and provide a foundation for the development of new type-specific diagnostic and therapeutic strategies for this disease.
MATERIALS AND METHODS
Establishment of USPC and NEC primary cell lines
A total of 15 primary cell lines (i.e., 10 USPC and five NEC) were established after sterile processing of samples from surgical biopsies collected between 1997 and 2004 at the University of Arkansas for Medical Sciences, as previously described for USPC specimens (Santin et al, 2002) and NEC cultures (Bongso et al, 1988). All fresh samples were obtained with appropriate consent according to IRB guidelines. Tumours were staged according to the FIGO operative staging system. A total abdominal hysterectomy with bilateral salpingo oophorectomy and bilateral pelvic lymphadenectomy was performed in all uterine carcinoma patients, while normal endometrial tissue was obtained from consenting donors undergoing surgery for benign pathology. No patient received chemotherapy or radiation before surgery. The patient characteristics are described in Table 1. Tumour cells were collected for RNA extraction at a confluence of 50–80% after a minimum of two to a maximum of 10 passages in vitro. The epithelial nature and the purity of USPC and NEC cultures were verified by immunohistochemical staining and flow-cytometric analysis with antibodies against cytokeratin and vimentin as described previously (Bongso et al, 1988; Santin et al, 2002). Only primary cultures which had at least 90% viability and contained >99% epithelial cells were used for total RNA extraction.
Table 1. Characteristics of the patients.
| Patient | Age | Race | Stage |
|---|---|---|---|
| USPC 1 | 65 | White | IV B |
| USPC 2 | 75 | Afro-American | III C |
| USPC 3 | 75 | Afro-American | IV A |
| USPC 4 | 59 | White | IV A |
| USPC 5 | 59 | White | III C |
| USPC 6 | 62 | Afro-American | IV B |
| USPC 7 | 63 | Afro-American | III C |
| USPC 8 | 61 | Afro-American | III C |
| USPC 9 | 78 | White | III C |
| USPC 10 | 64 | Afro-American | IV A |
RNA purification and microarray hybridization and analysis
Detailed protocols for RNA purification, cDNA synthesis, cRNA preparation, and hybridization to the Affymetrix Human U95Av2 GeneChip microarray were performed according to the manufacturer's protocols, as reported previously (Zhan et al, 2002).
Data processing
All data used in our analyses were derived from Affymetrix 5.0 software. GeneChip 5.0 output files are given as a signal that represents the difference between the intensities of the sequence-specific perfect match probe set and the mismatch probe set, or as a detection of present, marginal, or absent signals as determined by the GeneChip 5.0 algorithm. Gene arrays were scaled to an average signal of 1500 and then analysed independently. Signal calls were transformed by the log base 2 and each sample was normalized to give a mean of 0 and variance of 1.
Gene expression data analysis
Statistical analyses of the data were performed with the software packages SPSS10.0 (SPSS, Chicago, IL) and the significance analysis of microarrays (SAM) method (Tusher et al, 2001). Genes were selected for analysis based on detection and fold change. In each comparison, genes having ‘present’ detection calls in more than half of the samples in the overexpressed gene group were retained for statistical analysis if they showed >5-fold change between groups. Retained genes were subjected to SAM to establish a false discovery rate (FDR), then further filtered via the Wilcoxon rank sum (WRS) test at alpha=0.05. The FDR obtained from the initial SAM analysis was assumed to characterise genes found significant via WRS.
Gene cluster/treeview
The hierarchical clustering of average-linkage method with the centred correlation metric was used (Eisen et al, 1998). For the unsupervised hierarchical clustering, a total of 7328 probe sets were scanned across 10 USPCs and five NECs. The 7328 probe sets were derived from 12 588 by filtering out all control genes, all genes with absent detections, and genes not fulfilling the test of standard deviation greater than 0.5 (0.5 being the log base 2 of the signal). Only genes significantly expressed by both WRS and SAM analyses and whose average change in expression level was at least five-fold are shown in the Results section.
Quantitative real-time PCR (q-RT–PCR)
Quantitative real-time PCR was performed with an ABI Prism 7000 Sequence Analyzer using the manufacturer's recommended protocol (Applied Biosystems, Foster City, CA, USA) to validate differential expression of selected genes in samples from all 15 primary cell lines (10 USPC and five NEC). Each reaction was run in triplicate. The comparative threshold cycle (CT) method was used for the calculation of amplification fold as specified by the manufacturer. Briefly, 5 μg of total RNA from each sample was reverse transcribed using SuperScript II Rnase H Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). In all, 10 μl of reverse-transcribed RNA samples (from 500 μl of total volume) was amplified by using the TaqMan Universal PCR Master Mix (Applied Biosystems) to produce PCR products specific for cyclin-dependent kinase inhibitor 2A (CDKN2A/p16 and CDKN2A/p14ARF), L1 cell adhesion molecule (L1CAM), claudin-3, claudin-4, GRB7, and c-erbB2. Primers specific for 18s ribosomal RNA and empirically determined ratios of 18s competimers (Applied Biosystems) were used to control for the amounts of cDNA generated from each sample. Sequences for primers and probes are available on request. Differences among USPC and NEC in the q-RT–PCR expression data were tested using the Kruskal–Wallis nonparametric test. Pearson product–moment correlations were used to estimate the degree of association between the microarray and q-RT–PCR data.
Claudin-4 immunostaining of formalin-fixed tumour tissues
Claudin-4 protein expression was evaluated by immunohistochemical staining on formalin-fixed tumour tissue from which primary cultures were obtained. In addition, to further confirm transcriptional profiling results of USPC, claudin-4 marker was also evaluated by immunohistochemistry in a second independent set of eight USPC clinical tissue samples obtained from patients harbouring advanced stage disease (i.e., stage III and IV) treated at the UAMS during the same period. Study blocks were selected after histopathologic review by a surgical pathologist. The most representative haematoxylin and eosin-stained block sections were used for each specimen. Briefly, immunohistochemical stains were performed on 4-μm-thick sections of formalin-fixed, paraffin-embedded tissue. After pretreatment with 10 mM citrate buffer at pH 6.0 using a steamer, they were incubated with mouse anti-claudin-4 antibodies (cat. #: 18-7341; Zymed Laboratories Inc., San Francisco, CA, USA) at 1 : 2000 dilution. Antigen-bound primary antibodies were detected using standard avidin–biotin immunoperoxidase complex (Dako Corp., Carpinteria, CA, USA). Cases with less than 10% staining in tumour cells were considered negative for claudin expression, while positive cases were classified as follows regarding the intensity of claudin-4 protein expression: (a) +, focal membrane staining; (b) ++, diffuse membrane staining; and (c) +++, diffuse membrane and cytoplasmic staining.
Clostridium perfringens enterotoxin (CPE) treatment of primary USPC cell lines and trypan blue exclusion test
Tumour samples obtained from three patients harbouring advanced USPC (i.e., USPC 1, USPC 2, and USPC 3) and two NEC cultures derived from similar-aged women were seeded at a concentration of 1 × 105 cells well−1 into six-well culture plates (Costar, Cambridge, MA, USA) with the appropriate medium. Tumour samples and control cell lines were grown to 80% confluence. After washing and renewal of the medium, recombinant CPE cloned and purified as previously described (Michl et al, 2001) was added to final concentrations ranging from 0.03 to 3.3 μg ml−1. After incubation for 60 min to 24 h at 37°C, 5% CO2, floating cells were removed and stored, and attached cells were trypsinised and pooled with the floating cells. After staining with trypan blue, viability was determined by counting the number of trypan blue-positive cells and the total cell number.
RESULTS
Gene expression profiles distinguish USPC from NEC and identify differentially expressed genes
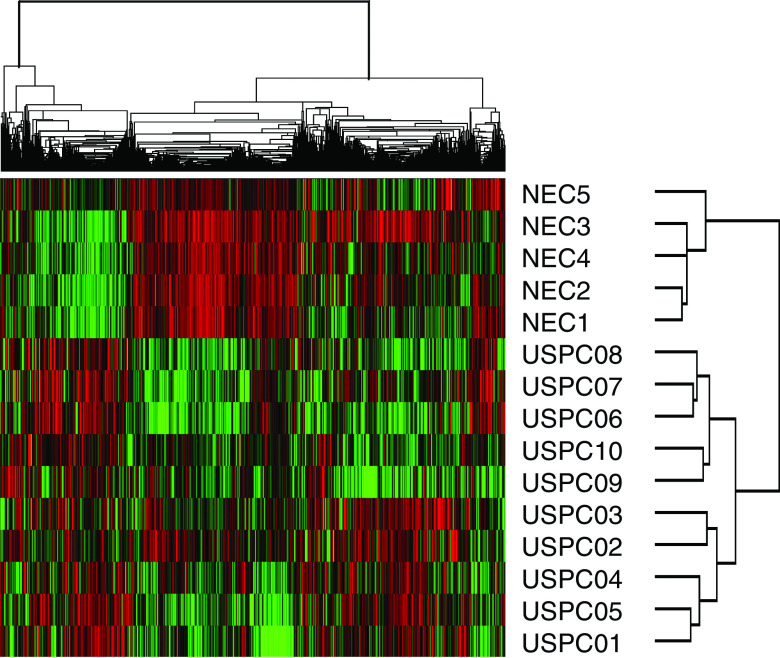
To minimise the risk of contamination of USPC RNA with that of normal cells or tumour cells with different histology (i.e., endometrioid or clear cells), as well as to reduce the complexity of gene expression data analysis, in this study we extracted RNA from short-term primary tumour cell cultures collected only from USPC with single-type differentiation (i.e., pure USPC). Short-term USPC and NEC cell cultures, minimising the risk of a selection bias inherent in any long-term in vitro growth, may provide an opportunity to study differential gene expression between highly enriched populations of normal and tumour-derived epithelial cells. Accordingly, comprehensive gene expression profiles of 10 primary USPC and five primary NEC cell lines were generated using high-density oligonucleotide arrays with 12 588 probe sets, which in total interrogated some 10 000 genes. Using unsupervised hierarchical cluster analysis with 7238 probe sets, we identified differences in gene expression between USPC and NEC, which readily distinguished the two groups of primary cultures. As shown in Figure 1, all 10 USPC were found to group together in the rightmost columns of the dendrogram. Similarly, in the leftmost columns, all five NEC were found to cluster tightly together. After filtering out most ‘absent’ genes, the SAM and the nonparametric WRS test (P<0.05) were performed to identify genes differentially expressed between USPC and NEC. A total of 2829 probe sets were found differentially expressed between USPC and NEC with P<0.05 by WRS and with a median FDR of 0.35% and a 90th percentile FDR of 0.59% by SAM. Of the 2829 aforementioned probe sets, there were 529 probe sets showing >5-fold change. As shown in Table 2, a group of 139 probe sets were found highly expressed in USPC and underexpressed in NEC. Included in this group of genes are CDKN2A/p16/p14ARF (101-fold), L1CAM (25-fold), claudin-3 (eight-fold), and claudin-4 (12-fold), kallikrein 6 (protease M) (19-fold), and kallikrein 10 (NES1) (23-fold), interleukin-6 (19-fold), interleukin-18 (10-fold), and plasminogen activator receptor (PLAUR) (seven-fold) (Table 2). Importantly, c-erbB2, a gene recently found by our group to be highly differentially expressed in USPC when compared to ovarian serous papillary tumours (Santin et al, 2004), was 14-fold more highly expressed in USPC than in NEC (Table 2). The second profile was represented by 390 genes that were highly expressed in NEC and underexpressed in USPC. Table 3 depicts the genes showing >10-fold change. Included in this group of genes are transforming growth factor beta receptor III, platelet-derived growth factor receptor alpha, SEMACAP3, ras homolog gene family, member I (ARHI), and differentially downregulated in ovarian carcinoma 1 (DOC1).
Figure 1.
Unsupervised hierarchical clustering of 15 primary uterine cell lines (i.e., 10 USPC and five NEC). The cluster is colour coded using red for upregulation, green for downregulation, and black for median expression. Agglomerative clustering of genes is illustrated with dendrograms.
Table 2. Upregulated genes expressed at least five-fold higher in USPC compared with NEC.
| Probe set | Gene symbol | SAM | P of WRS | Ratio USPC/NEC | Title |
|---|---|---|---|---|---|
| 1713_s_at | CDKN2A a | 10.592 | 0.0027 | 101.9077377 | ‘Cyclin-dependent kinase inhibitor 2A (melanoma, p16)’ |
| 36288_at | KRTHB1 | 4.573 | 0.0027 | 77.3983986 | ‘Keratin, hair, basic, 1’ |
| 33272_at | SAA1 | 3.778 | 0.0063 | 45.74937337 | Serum amyloid A1 |
| 41294_at | KRT7 | 7.173 | 0.0027 | 41.46788873 | Keratin 7 |
| 32154_at | TFAP2A | 7.637 | 0.0027 | 32.47929396 | Transcription factor AP-2 alpha |
| 31610_at | MAP17 | 3.621 | 0.0093 | 30.28302802 | Membrane-associated protein 17 |
| 408_at | — | 4.070 | 0.0063 | 30.14111158 | — |
| 32821_at | LCN2 | 5.126 | 0.0027 | 27.69975608 | Lipocalin 2 (oncogene 24p3) |
| 35174_i_at | EEF1A2 | 2.840 | 0.0278 | 26.80482891 | Elongation factor 1 alpha 2 |
| 38551_at | L1CAM | 3.115 | 0.0196 | 25.60938089 | ‘L1 cell adhesion molecule |
| 38249_at | VGLL1 | 5.274 | 0.0027 | 24.69495091 | Vestigial like 1 (Drosophila) |
| 35879_at | GAL | 5.594 | 0.0027 | 23.48953559 | Galanin |
| 36838_at | KLK10 | 3.455 | 0.0136 | 23.17518549 | Kallikrein 10 |
| 38299_at | IL6 | 3.630 | 0.0041 | 19.05873079 | ‘Interleukin 6 (interferon, beta 2)’ |
| 38051_at | MAL | 4.878 | 0.0041 | 17.51555106 | ‘mal, T-cell differentiation protein’ |
| 41469_at | PI3 | 2.854 | 0.0063 | 16.90464558 | ‘Protease inhibitor 3, skin-derived (SKALP)’ |
| 40412_at | PTTG1 | 5.218 | 0.0027 | 16.61222352 | Pituitary tumour-transforming 1 |
| 1886_at | WNT7A | 3.544 | 0.0196 | 16.11519778 | ‘Wingless-type MMTV integration site family, member 7A’ |
| 33128_s_at | CST6 | 4.222 | 0.0136 | 15.97856318 | Cystatin E/M |
| 38414_at | CDC20 | 7.317 | 0.0027 | 15.64601435 | CDC20 cell division cycle 20 homolog (S. cerevisiae) |
| 34012_at | KRTHA4 | 2.411 | 0.0278 | 15.37247475 | ‘Keratin, hair, acidic, 4’ |
| 37554_at | KLK6 | 3.785 | 0.0093 | 15.23781352 | ‘Kallikrein 6 (neurosin, zyme)’ |
| 1802_s_at | ERBB2 | 2.566 | 0.0136 | 14.52012028 | ‘v-erb-b2’ |
| 41060_at | CCNE1 | 6.092 | 0.0027 | 14.16647068 | Cyclin E1 |
| 36837_at | KIF2C | 6.130 | 0.0027 | 14.1328483 | Kinesin family member 2C |
| 34213_at | KIBRA | 5.301 | 0.0027 | 13.27228177 | KIBRA protein |
| 1651_at | UBE2C | 5.554 | 0.0027 | 12.87617243 | Ubiquitin-conjugating enzyme E2C |
| 35276_at | CLDN4 | 6.381 | 0.0027 | 12.74825421 | Claudin 4 |
| 36990_at | UCHL1 | 4.623 | 0.0027 | 12.30505908 | Ubiquitin carboxyl-terminal esterase L1 |
| 35977_at | DKK1 | 4.495 | 0.0041 | 12.25382636 | Dickkopf homolog 1 (Xenopus laevis) |
| 36113_s_at | TNNT1 | 4.072 | 0.0027 | 11.93824813 | ‘Troponin T1, skeletal, slow’ |
| 2011_s_at | BIK | 3.451 | 0.0063 | 11.66959681 | BCL2-interacting killer (apoptosis-inducing) |
| 543_g_at | CRABP1 | 3.193 | 0.0093 | 11.55494382 | Cellular retinoic acid binding protein 1 |
| 34852_g_at | STK6 | 6.225 | 0.0027 | 11.51812047 | Serine/threonine kinase 6 |
| 33483_at | NMU | 4.094 | 0.0027 | 11.42057993 | Neuromedin U |
| 39109_at | TPX2 | 6.162 | 0.0027 | 11.29208457 | ‘TPX2, microtubule-associated protein homologue’ |
| 37018_at | HIST1H1C | 2.270 | 0.0278 | 10.74270622 | ‘Histone 1, H1c’ |
| 1165_at | IL18 | 3.221 | 0.0041 | 10.65596528 | Interleukin 18 (interferon-gamma-inducing factor) |
| 36477_at | TNNI3 | 2.867 | 0.0136 | 10.61101382 | ‘Troponin I, cardiac’ |
| 572_at | TTK | 3.720 | 0.0093 | 9.723902052 | TTK protein kinase |
| 31542_at | FLG | 2.622 | 0.0196 | 9.600831601 | Filaggrin |
| 35937_at | MICB | 4.238 | 0.0093 | 9.460109582 | MHC class I polypeptide-related sequence B |
| 36155_at | SPOCK2 | 2.278 | 0.0278 | 9.216570003 | ‘Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2’ |
| 32186_at | SLC7A5 | 4.149 | 0.0063 | 9.121679665 | ‘Solute carrier family 7 (cationic amino-acid transporter, y+ system), member 5’ |
| 35766_at | KRT18 | 5.933 | 0.0027 | 9.01220054 | Keratin 18 |
| 35822_at | BF | 3.267 | 0.0063 | 8.952514469 | ‘B-factor, properdin’ |
| 35714_at | PDXK | 6.550 | 0.0027 | 8.898191704 | ‘Pyridoxal (pyridoxine, vitamin B6) kinase’ |
| 1369_s_at | — | 2.679 | 0.0196 | 8.773380878 | — |
| 40079_at | RAI3 | 4.516 | 0.0063 | 8.626843209 | Retinoic acid induced 3 |
| 37168_at | LAMP3 | 2.838 | 0.0136 | 8.616807346 | Lysosomal-associated membrane protein 3 |
| 39704_s_at | HMGA1 | 5.322 | 0.0027 | 8.597894471 | High-mobility group AT-hook 1 |
| 1887_g_at | WNT7A | 3.003 | 0.0196 | 8.491813649 | ‘Wingless-type MMTV integration site family, member 7A’ |
| 36929_at | LAMB3 | 5.770 | 0.0027 | 8.354149098 | ‘Laminin, beta 3’ |
| 527_at | CENPA | 6.126 | 0.0027 | 8.32992789 | ‘Centromere protein A, 17 kDa’ |
| 41081_at | BUB1 | 4.883 | 0.0027 | 8.213759056 | BUB1 budding uninhibited by benzimidazoles 1 homologue |
| 885_g_at | ITGA3 | 4.447 | 0.0027 | 8.20660555 | ‘Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)’ |
| 2021_s_at | CCNE1 | 4.399 | 0.0041 | 8.199388463 | Cyclin E1 |
| 33904_at | CLDN3 | 3.296 | 0.0136 | 8.020010794 | Claudin 3 |
| 33730_at | RAI3 | 4.648 | 0.0041 | 7.923899439 | Retinoic acid induced 3 |
| 34736_at | CCNB1 | 5.078 | 0.0063 | 7.896644626 | Cyclin B1 |
| 757_at | ANXA2 | 3.514 | 0.0063 | 7.870466864 | Annexin A2 |
| 910_at | TK1 | 3.934 | 0.0093 | 7.869091533 | ‘Thymidine kinase 1, soluble’ |
| 34851_at | STK6 | 4.491 | 0.0041 | 7.764803777 | Serine/threonine kinase 6 |
| 34703_f_at | — | 2.275 | 0.0278 | 7.710260816 | — |
| 34715_at | FOXM1 | 5.318 | 0.0027 | 7.659023602 | Forkhead box M1 |
| 38971_r_at | TNIP1 | 6.800 | 0.0027 | 7.595036872 | TNFAIP3 interacting protein 1 |
| 32263_at | CCNB2 | 4.246 | 0.0063 | 7.578513543 | Cyclin B2 |
| 1680_at | GRB7 | 4.013 | 0.0027 | 7.471384928 | Growth factor receptor-bound protein 7 |
| 38247_at | F2RL1 | 3.185 | 0.0093 | 7.432476326 | Coagulation factor II (thrombin) receptor-like 1 |
| 160025_at | TGFA | 6.088 | 0.0027 | 7.355344272 | ‘Transforming growth factor, alpha’ |
| 1945_at | CCNB1 | 5.298 | 0.0041 | 7.291039832 | Cyclin B1 |
| 31792_at | ANXA3 | 4.873 | 0.0041 | 7.266892828 | Annexin A3 |
| 182_at | ITPR3 | 5.432 | 0.0027 | 7.172450367 | ‘Inositol 1,4,5-triphosphate receptor, type 3’ |
| 1117_at | CDA | 2.937 | 0.0093 | 7.114518646 | Cytidine deaminase |
| 902_at | EPHB2 | 5.186 | 0.0027 | 7.065363569 | EphB2 |
| 634_at | PRSS8 | 5.216 | 0.0041 | 7.001894703 | ‘Protease, serine, 8 (prostasin)’ |
| 41169_at | PLAUR | 3.982 | 0.0063 | 7.00139089 | ‘Plasminogen activator, urokinase receptor’ |
| 33203_s_at | FOXD1 | 3.464 | 0.0093 | 6.989749222 | Forkhead box D1 |
| 40095_at | CA2 | 4.285 | 0.0027 | 6.946396937 | Carbonic anhydrase II |
| 38940_at | AD024 | 5.065 | 0.0041 | 6.928406028 | AD024 protein |
| 34348_at | SPINT2 | 6.263 | 0.0027 | 6.877224695 | ‘Serine protease inhibitor, Kunitz type, 2’ |
| 33933_at | WFDC2 | 3.344 | 0.0136 | 6.820073691 | WAP four-disulphide core domain 2 |
| 35281_at | LAMC2 | 3.347 | 0.0093 | 6.7580474 | ‘Laminin, gamma 2’ |
| 349_g_at | KIFC1 | 5.275 | 0.0041 | 6.700913018 | Kinesin family member C1 |
| 33218_at | ERBB2 | 2.710 | 0.0027 | 6.615105998 | ‘v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, neuro/glioblastoma derived oncogene homolog (avian)’ |
| 38881_i_at | TRIM16 | 3.001 | 0.0196 | 6.506893575 | Tripartite motif-containing 16 |
| 1536_at | CDC6 | 4.666 | 0.0041 | 6.463305623 | CDC6 cell division cycle 6 homolog (S. cerevisiae) |
| 38482_at | CLDN7 | 4.931 | 0.0041 | 6.409117877 | Claudin 7 |
| 40697_at | CCNA2 | 3.396 | 0.0093 | 6.40768505 | Cyclin A2 |
| 41688_at | TM4SF11 | 4.390 | 0.0027 | 6.366861533 | Transmembrane 4 superfamily member 11 (plasmolipin) |
| 38158_at | ESPL1 | 6.007 | 0.0027 | 6.225688779 | Extra spindle poles like 1 (S. cerevisiae) |
| 38474_at | CBS | 3.380 | 0.0093 | 6.212078913 | Cystathionine-beta-synthase |
| 36483_at | GALNT3 | 3.890 | 0.0041 | 6.181109111 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| 35372_r_at | IL8 | 2.360 | 0.0278 | 6.133149591 | Interleukin 8 |
| 41585_at | KIAA0746 | 4.436 | 0.0027 | 6.092207586 | KIAA0746 protein |
| 36832_at | B3GNT3 | 5.457 | 0.0027 | 5.941291793 | ‘UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3’ |
| 1107_s_at | G1P2 | 3.938 | 0.0063 | 5.923287019 | ‘Interferon, alpha-inducible protein (clone IFI-15K)’ |
| 35207_at | SCNN1A | 3.076 | 0.0136 | 5.920739634 | ‘Sodium channel, nonvoltage-gated 1 alpha’ |
| 36863_at | HMMR | 2.830 | 0.0196 | 5.905038013 | Hyaluronan-mediated motility receptor (RHAMM) |
| 38631_at | TNFAIP2 | 4.924 | 0.0027 | 5.897745642 | ‘Tumour necrosis factor, alpha-induced protein 2’ |
| 36813_at | TRIP13 | 5.666 | 0.0027 | 5.870351247 | Thyroid hormone receptor interactor 13 |
| 41048_at | PMAIP1 | 3.490 | 0.0062 | 5.853172336 | Phorbol-12-myristate-13-acetate-induced protein 1 |
| 2084_s_at | ETV4 | 3.743 | 0.0093 | 5.798002338 | ‘ets variant gene 4 (E1A enhancer binding protein, E1AF)’ |
| 33245_at | MAPK13 | 3.775 | 0.0136 | 5.766618762 | Mitogen-activated protein kinase 13 |
| 37347_at | CKS1B | 5.543 | 0.0027 | 5.762817533 | CDC28 protein kinase regulatory subunit 1B |
| 34282_at | NFE2L3 | 2.668 | 0.0136 | 5.734907375 | Nuclear factor (erythroid-derived 2)-like 3 |
| 330_s_at | — | 4.026 | 0.0041 | 5.726752495 | — |
| 41732_at | na | 6.920 | 0.0027 | 5.706487141 | Similar to My016 protein |
| 1516_g_at | — | 6.726 | 0.0027 | 5.63870137 | — |
| 904_s_at | TOP2A | 3.419 | 0.0063 | 5.634251452 | Topoisomerase (DNA) II alpha 170 kDa |
| 36041_at | EXO1 | 4.971 | 0.0027 | 5.59235892 | Exonuclease 1 |
| 33143_s_at | SLC16A3 | 4.007 | 0.0063 | 5.56591457 | ‘Solute carrier family 16 (monocarboxylic acid transporters), member 3’ |
| 37228_at | PLK | 4.501 | 0.0041 | 5.564532365 | Polo-like kinase (Drosophila) |
| 1854_at | MYBL2 | 4.117 | 0.0063 | 5.54317592 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 |
| 40407_at | KPNA2 | 4.189 | 0.0041 | 5.51635645 | ‘Karyopherin alpha 2 (RAG cohort 1, importin alpha 1)’ |
| 33282_at | LAD1 | 3.904 | 0.0063 | 5.509367036 | Ladinin 1 |
| 40145_at | TOP2A | 3.307 | 0.0093 | 5.48127065 | Topoisomerase (DNA) II alpha 170 kDa |
| 1100_at | IRAK1 | 5.530 | 0.0027 | 5.470162749 | Interleukin-1 receptor-associated kinase 1 |
| 37883_i_at | AF038169 | 3.160 | 0.0027 | 5.460495655 | Hypothetical protein AF038169 |
| 37343_at | ITPR3 | 5.258 | 0.0027 | 5.449013729 | ‘Inositol 1,4,5-triphosphate receptor, type 3’ |
| 31598_s_at | GALE | 4.647 | 0.0027 | 5.442955253 | ‘Galactose-4-epimerase, UDP-’ |
| 889_at | ITGB8 | 2.744 | 0.0093 | 5.370592815 | ‘Integrin, beta 8’ |
| 37558_at | IMP-3 | 3.123 | 0.0093 | 5.364127468 | IGF-II mRNA-binding protein 3 |
| 32715_at | VAMP8 | 5.686 | 0.0027 | 5.352873419 | Vesicle-associated membrane protein 8 (endobrevin) |
| 36312_at | SERPINB8 | 3.611 | 0.0027 | 5.327343554 | ‘Serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 8’ |
| 37210_at | INA | 3.551 | 0.0063 | 5.307526088 | ‘Internexin neuronal intermediate filament protein, alpha’ |
| 35699_at | BUB1B | 3.665 | 0.0196 | 5.279075308 | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) |
| 32787_at | ERBB3 | 2.658 | 0.0041 | 5.247404657 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) |
| 32275_at | SLPI | 3.726 | 0.0041 | 5.221163981 | Secretory leukocyte protease inhibitor (antileukoproteinase) |
| 893_at | E2-EPF | 3.775 | 0.0063 | 5.196412396 | Ubiquitin carrier protein |
| 41583_at | FEN1 | 5.481 | 0.0027 | 5.196005796 | Flap structure-specific endonuclease 1 |
| 41781_at | PPFIA1 | 4.113 | 0.0027 | 5.194931774 | ‘Protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1’ |
| 40726_at | KIF11 | 2.941 | 0.0093 | 5.1806793 | Kinesin family member 11 |
| 41400_at | TK1 | 4.246 | 0.0093 | 5.167172588 | ‘Thymidine kinase 1, soluble’ |
| 41409_at | C1orf38 | 3.109 | 0.0063 | 5.100239097 | Chromosome 1 open reading frame 38 |
| 40425_at | EFNA1 | 2.738 | 0.0196 | 5.067718102 | Ephrin-A1 |
| 32081_at | CIT | 6.162 | 0.0027 | 5.043567722 | ‘Citron (rho-interacting, serine/threonine kinase 21)’ |
| 1108_s_at | EPHA1 | 4.864 | 0.0027 | 5.040980858 | EphA1 |
| 33338_at | STAT1 | 3.275 | 0.0063 | 5.029498048 | ‘Signal transducer and activator of transcription 1, 91 kDa’ |
Genes selected for further analysis are identified with bold letters.
Table 3. Upregulated genes expressed at least 10-fold higher in NEC compared with USPC.
| Probe set | Gene symbol | SAM | P of WRS | Ratio NEC/USPC | Title |
|---|---|---|---|---|---|
| 774_g_at | MYH11 | 17.611 | 0.0027 | 1014.968759 | ‘Myosin, heavy polypeptide 11, smooth muscle’ |
| 773_at | MYH11 | 14.265 | 0.0027 | 968.0223497 | ‘Myosin, heavy polypeptide 11, smooth muscle’ |
| 32582_at | MYH11 | 11.277 | 0.0027 | 212.2458648 | ‘Myosin, heavy polypeptide 11, smooth muscle’ |
| 36681_at | APOD | 11.714 | 0.0027 | 137.3140116 | Apolipoprotein D |
| 1501_at | IGF1 | 8.321 | 0.0027 | 128.0651104 | Insulin-like growth factor 1 (somatomedin C) |
| 39325_at | EBAF | 8.177 | 0.0027 | 123.064609 | ‘Endometrial bleeding associated factor)’ |
| 767_at | — | 12.707 | 0.0027 | 119.5409103 | — |
| 40398_s_at | MEOX2 | 10.182 | 0.0027 | 114.9489897 | Mesenchyme homeo box 2 |
| 40776_at | DES | 9.738 | 0.0027 | 109.416397 | Desmin |
| 1197_at | ACTG2 | 5.861 | 0.0027 | 96.65715109 | ‘Actin, gamma 2, smooth muscle, enteric’ |
| 36627_at | SPARCL1 | 7.724 | 0.0027 | 93.89302842 | ‘SPARC-like 1 (mast9, hevin)’ |
| 37407_s_at | MYH11 | 11.893 | 0.0027 | 88.70362054 | ‘Myosin, heavy polypeptide 11, smooth muscle’ |
| 39673_i_at | ECM2 | 8.459 | 0.0027 | 82.52353674 | ‘Extracellular matrix protein 2, female organ and adipocyte specific’ |
| 39701_at | PEG3 | 7.645 | 0.0027 | 73.87997223 | Paternally expressed gene 3 |
| 39066_at | MFAP4 | 9.383 | 0.0027 | 68.84723295 | Microfibrillar-associated protein 4 |
| 38737_at | IGF1 | 6.341 | 0.0027 | 65.13666715 | Insulin-like growth factor 1 (somatomedin C) |
| 35730_at | ADH1B | 9.759 | 0.0027 | 64.94146631 | ‘Alcohol dehydrogenase IB (class I), beta polypeptide’ |
| 41124_r_at | ENPP2 | 7.007 | 0.0027 | 53.43118221 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) |
| 36749_at | CPA3 | 9.096 | 0.0027 | 53.07347991 | Carboxypeptidase A3 (mast cell) |
| 39616_at | PTGER3 | 10.188 | 0.0027 | 52.65326101 | Prostaglandin E receptor 3 (subtype EP3) |
| 33440_at | TCF8 | 7.220 | 0.0027 | 49.54168001 | Transcription factor 8 |
| 32666_at | CXCL12 | 7.168 | 0.0027 | 49.2359637 | Chemokine (C–X–C motif) ligand 12 |
| 38734_at | PLN | 9.182 | 0.0027 | 47.86730799 | Phospholamban |
| 34203_at | CNN1 | 5.051 | 0.0027 | 47.0209722 | ‘Calponin 1, basic, smooth muscle’ |
| 37576_at | PCP4 | 12.468 | 0.0027 | 45.45542357 | Purkinje cell protein 4 |
| 36834_at | MOXD1 | 9.736 | 0.0027 | 44.20917891 | ‘Monooxygenase, DBH-like 1’ |
| 37247_at | TCF21 | 9.810 | 0.0027 | 42.40051895 | Transcription factor 21 |
| 37701_at | RGS2 | 7.408 | 0.0027 | 42.29038464 | ‘Regulator of G-protein signalling 2, 24 kDa’ |
| 779_at | COL4A6 | 5.840 | 0.0027 | 41.96325597 | ‘Collagen, type IV, alpha 6’ |
| 37394_at | C7 | 9.835 | 0.0027 | 40.72326307 | Complement component 7 |
| 36533_at | PTGIS | 6.097 | 0.0027 | 39.10052985 | Prostaglandin I2 (prostacyclin) synthase |
| 32689_s_at | — | 6.178 | 0.0027 | 36.96897988 | — |
| 32905_s_at | TPSB2 | 6.873 | 0.0027 | 36.85974711 | Tryptase beta 2 |
| 33240_at | SEMACAP3 | 4.814 | 0.0027 | 36.54583796 | Likely ortholog of mouse semaF cytoplasmic domain associated protein 3 |
| 1466_s_at | FGF7 | 8.628 | 0.0027 | 35.60413415 | Fibroblast growth factor 7 |
| 32686_at | PTGER3 | 7.397 | 0.0027 | 35.31357002 | Prostaglandin E receptor 3 (subtype EP3) |
| 39690_at | ALP | 6.951 | 0.0027 | 33.76045029 | Alpha-actinin-2-associated LIM protein |
| 38427_at | COL15A1 | 7.196 | 0.0027 | 32.85391153 | ‘Collagen, type XV, alpha 1’ |
| 39939_at | COL4A6 | 6.846 | 0.0027 | 32.53815505 | ‘Collagen, type IV, alpha 6’ |
| 37630_at | NRLN1 | 5.896 | 0.0027 | 31.03030255 | Neuralin 1 |
| 35717_at | ABCA8 | 7.587 | 0.0027 | 30.63819827 | ‘ATP-binding cassette, subfamily A, member 8’ |
| 1709_g_at | MAPK10 | 6.522 | 0.0027 | 30.54331363 | Mitogen-activated protein kinase 10 |
| 35679_s_at | DPP6 | 5.899 | 0.0027 | 30.33959319 | Dipeptidylpeptidase 6 |
| 35740_at | EMILIN1 | 7.445 | 0.0027 | 29.45261895 | Elastin microfibril interfacer 1 |
| 41123_s_at | ENPP2 | 6.597 | 0.0027 | 29.29614059 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (autotaxin) |
| 755_at | ITPR1 | 8.211 | 0.0027 | 29.02124207 | ‘Inositol 1,4,5-triphosphate receptor, type 1’ |
| 32847_at | MYLK | 5.388 | 0.0027 | 28.22518538 | ‘Myosin, light polypeptide kinase’ |
| 38001_at | CACNA1C | 5.740 | 0.0027 | 27.31347302 | ‘Calcium channel, alpha 1C subunit’ |
| 37279_at | GEM | 9.925 | 0.0027 | 26.56613192 | GTP-binding protein overexpressed in skeletal muscle |
| 36396_at | — | 5.901 | 0.0027 | 26.32457357 | Homo sapiens mRNA; cDNA DKFZp586N2020 (from clone DKFZp586N2020) |
| 41137_at | PPP1R12B | 12.043 | 0.0027 | 25.97089935 | ‘Protein phosphatase 1, regulatory subunit 12B’ |
| 41405_at | SFRP4 | 10.077 | 0.0027 | 23.90433923 | Secreted frizzled-related protein 4 |
| 40775_at | ITM2A | 6.152 | 0.0027 | 23.83083993 | Integral membrane protein 2A |
| 38059_g_at | DPT | 6.508 | 0.0027 | 23.76376781 | Dermatopontin |
| 41504_s_at | MAF | 5.331 | 0.0027 | 23.65323618 | v-maf |
| 1596_g_at | TEK | 4.542 | 0.0027 | 23.37422615 | ‘TEK tyrosine kinase, endothelial (venous malformations, multiple cutaneous and mucosal)’ |
| 914_g_at | ERG | 6.105 | 0.0027 | 22.63829292 | v-ets erythroblastosis virus E26 oncogene like |
| 34283_at | — | 5.390 | 0.0041 | 22.19917455 | ‘Homo sapiens, clone IMAGE:4791553, mRNA’ |
| 34388_at | COL14A1 | 9.452 | 0.0027 | 21.48165459 | ‘Collagen, type XIV, alpha 1 (undulin)’ |
| 38994_at | SOCS2 | 6.600 | 0.0027 | 21.47663968 | Suppressor of cytokine signaling 2 |
| 36065_at | LDB2 | 5.790 | 0.0027 | 21.19321308 | LIM domain binding 2 |
| 40230_at | FRZB | 5.962 | 0.0027 | 21.00467856 | Frizzled-related protein |
| 33790_at | CCL15 | 9.017 | 0.0027 | 20.5510028 | Chemokine (C–C motif) ligand 15 |
| 33890_at | RGS5 | 7.347 | 0.0027 | 20.2203337 | Regulator of G-protein signalling 5 |
| 36513_at | MAGP2 | 4.385 | 0.0041 | 20.10734871 | Microfibril-associated glycoprotein-2 |
| 32526_at | JAM3 | 6.129 | 0.0027 | 19.89641568 | Junctional adhesion molecule 3 |
| 32687_s_at | PTGER3 | 5.902 | 0.0027 | 19.7194642 | Prostaglandin E receptor 3 (subtype EP3) |
| 35638_at | CBFA2T1 | 10.470 | 0.0027 | 19.57233853 | ‘Core-binding factor, runt domain, alpha subunit 2; translocated to, 1; cyclin D-related’ |
| 34637_f_at | ADH1A | 6.291 | 0.0027 | 19.20847669 | ‘Alcohol dehydrogenase 1A (class I)’ |
| 34675_at | SBLF | 5.400 | 0.0027 | 19.04463285 | Stoned B-like factor |
| 38351_at | — | 5.899 | 0.0026 | 18.69131208 | Homo sapiens mRNA; cDNA DKFZp586L0120 (from clone DKFZp586L0120) |
| 1182_at | PLCL1 | 4.047 | 0.0027 | 18.50958764 | Phospholipase C-like 1 |
| 39681_at | ZNF145 | 6.266 | 0.0027 | 18.41823634 | ‘Zinc-finger protein 145 (Kruppel-like, expressed in promyelocytic leukemia)’ |
| 1708_at | MAPK10 | 6.336 | 0.0027 | 18.31250756 | Mitogen-activated protein kinase 10 |
| 37765_at | LMOD1 | 7.586 | 0.0027 | 18.11624265 | Leiomodin 1 (smooth muscle) |
| 1678_g_at | IGFBP5 | 4.237 | 0.0041 | 17.88349387 | Insulin-like growth factor binding protein 5 |
| 35358_at | TENC1 | 9.243 | 0.0027 | 17.68277053 | Tensin-like C1 domain-containing phosphatase |
| 33442_at | KIAA0367 | 6.830 | 0.0027 | 17.58195512 | KIAA0367 protein |
| 37249_at | PDE8B | 5.035 | 0.0027 | 17.25176904 | Phosphodiesterase 8B |
| 33834_at | CXCL12 | 7.736 | 0.0027 | 17.10893345 | Chemokine (C–X–C motif) ligand 12 (stromal cell-derived factor 1) |
| 35324_at | SLIT3 | 5.157 | 0.0027 | 17.08441711 | Slit homolog 3 (Drosophila) |
| 37015_at | ALDH1A1 | 4.941 | 0.0027 | 16.83181709 | ‘Aldehyde dehydrogenase 1 family, member A1’ |
| 39266_at | — | 6.237 | 0.0027 | 16.80733058 | Homo sapiens clone 24405 mRNA sequence |
| 33462_at | GPR105 | 4.907 | 0.0027 | 16.49634139 | G protein-coupled receptor 105 |
| 32488_at | COL3A1 | 3.928 | 0.0027 | 16.44993439 | ‘Collagen, type III, alpha 1 (Ehlers-Danlos syndrome type IV, autosomal dominant)’ |
| 483_g_at | CDH13 | 3.755 | 0.0027 | 16.43857351 | ‘Cadherin 13, H-cadherin (heart)’ |
| 38026_at | FBLN1 | 5.350 | 0.0027 | 16.06497318 | Fibulin 1 |
| 1909_at | BCL2 | 7.356 | 0.0027 | 16.01160367 | B-cell CLL/lymphoma 2 |
| 36245_at | HTR2B | 4.698 | 0.0027 | 15.77119379 | 5-Hydroxytryptamine (serotonin) receptor 2B |
| 32057_at | LRRC17 | 5.470 | 0.0027 | 15.6891271 | Leucine-rich repeat containing 17 |
| 39544_at | DMN | 5.361 | 0.0027 | 15.66465434 | Desmuslin |
| 37112_at | C6orf32 | 4.720 | 0.0027 | 15.65975413 | Chromosome 6 open reading frame 32 |
| 36733_at | FLJ32389 | 4.442 | 0.0027 | 15.64273348 | Hypothetical protein FLJ32389 |
| 1319_at | DDR2 | 5.400 | 0.0027 | 15.5817123 | ‘Discoidin domain receptor family, member 2’ |
| 38057_at | DPT | 8.479 | 0.0027 | 15.51358362 | Dermatopontin |
| 40358_at | GLI3 | 6.650 | 0.0027 | 15.11249492 | GLI-Kruppel family member GLI3 (Greig cephalopolysyndactyly syndrome) |
| 38627_at | HLF | 4.947 | 0.0027 | 14.89249894 | Hepatic leukemia factor |
| 1731_at | PDGFRA | 7.050 | 0.0027 | 14.84422948 | ‘Platelet-derived growth factor receptor, alpha polypeptide’ |
| 31897_at | DOC1 | 4.664 | 0.0027 | 14.80084961 | Downregulated in ovarian cancer 1 |
| 2073_s_at | CDH13 | 5.076 | 0.0027 | 14.79118507 | ‘Cadherin 13, H-cadherin (heart)’ |
| 38577_at | — | 7.283 | 0.0027 | 14.78670234 | ‘Clone DT1P1B6 mRNA, CAG repeat region’ |
| 38004_at | CSPG4 | 6.072 | 0.0027 | 14.40637488 | Chondroitin sulphate proteoglycan 4 |
| 32109_at | FXYD1 | 7.333 | 0.0027 | 14.33463902 | Phospholemman |
| 34853_at | FLRT2 | 9.834 | 0.0027 | 14.33080432 | Fibronectin leucine-rich transmembrane protein 2 |
| 38298_at | KCNMB1 | 7.046 | 0.0027 | 14.25816751 | ‘Potassium large-conductance calcium-activated channel, subfamily M, beta member 1’ |
| 41245_at | GDF10 | 6.643 | 0.0027 | 14.21467179 | Growth differentiation factor 10 |
| 38322_at | GAGEC1 | 6.215 | 0.0027 | 14.10508119 | ‘G antigen, family C, 1’ |
| 1198_at | EDNRB | 4.899 | 0.0027 | 14.08196951 | Endothelin receptor type B |
| 41505_r_at | MAF | 4.528 | 0.0027 | 14.0059518 | v-maf |
| 36976_at | CDH11 | 3.692 | 0.0027 | 14.00329993 | ‘Cadherin 11, type 2, OB-cadherin (osteoblast)’ |
| 32688_at | PTGER3 | 4.998 | 0.0027 | 13.68791213 | Prostaglandin E receptor 3 (subtype EP3) |
| 1575_at | ABCB1 | 6.210 | 0.0027 | 13.64196645 | ‘ATP-binding cassette, subfamily B’ |
| 32778_at | ITPR1 | 4.914 | 0.0027 | 13.60359986 | ‘Inositol 1,4,5-triphosphate receptor, type 1’ |
| 40737_at | KCNMA1 | 5.642 | 0.0027 | 13.60011778 | ‘potassium large-conductance calcium-activated channel, subfamily M, alpha member 1’ |
| 36569_at | TNA | 11.512 | 0.0027 | 13.14186685 | Tetranectin (plasminogen-binding protein) |
| 1897_at | TGFBR3 | 4.922 | 0.0041 | 12.99722509 | ‘Transforming growth factor, beta receptor III (betaglycan, 300 kDa)’ |
| 2087_s_at | CDH11 | 4.345 | 0.0027 | 12.96131894 | ‘Cadherin 11, type 2, OB-cadherin (osteoblast)’ |
| 34820_at | PTN | 4.413 | 0.0041 | 12.94626537 | ‘Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1)’ |
| 743_at | NAP1L3 | 7.859 | 0.0027 | 12.91061134 | Nucleosome assembly protein 1-like 3 |
| 1507_s_at | EDNRA | 4.319 | 0.0041 | 12.88466839 | Endothelin receptor type A |
| 38995_at | CLDN5 | 12.156 | 0.0027 | 12.86636376 | Claudin 5 (transmembrane protein deleted in velocardiofacial syndrome) |
| 1147_at | — | 7.084 | 0.0027 | 12.69931721 | — |
| 1954_at | KDR | 6.077 | 0.0027 | 12.6174344 | Kinase insert domain receptor (a type III receptor tyrosine kinase) |
| 38786_at | — | 5.676 | 0.0027 | 12.61170471 | Homo sapiens mRNA full-length insert cDNA clone EUROIMAGE 248114 |
| 40757_at | GZMA | 4.575 | 0.0041 | 12.56281091 | ‘Granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)’ |
| 36695_at | na | 5.776 | 0.0027 | 12.53905775 | Similar to hypothetical protein 6720478C22 |
| 32052_at | HBB | 5.107 | 0.0063 | 12.52262761 | ‘Haemoglobin, beta’ |
| 37446_at | GASP | 8.227 | 0.0027 | 12.39360585 | G protein-coupled receptor-associated protein |
| 38038_at | LUM | 4.462 | 0.0027 | 12.34757673 | Lumican |
| 32889_at | RPIB9 | 6.337 | 0.0027 | 12.34540513 | Rap2-binding protein 9 |
| 35234_at | RECK | 4.476 | 0.0027 | 12.33793353 | Reversion-inducing-cysteine-rich protein |
| 661_at | GAS1 | 4.648 | 0.0027 | 12.32556384 | Growth arrest-specific 1 |
| 41195_at | LPP | 6.292 | 0.0027 | 12.26636801 | LIM domain containing preferred translocation partner in lipoma |
| 32664_at | RNASE4 | 6.395 | 0.0027 | 11.89680804 | ‘Ribonuclease, RNase A family, 4’ |
| 39038_at | FBLN5 | 4.325 | 0.0027 | 11.89440528 | Fibulin 5 |
| 40693_at | KCNB1 | 4.952 | 0.0041 | 11.84773944 | ‘Potassium voltage-gated channel, Shab-related subfamily, member 1’ |
| 38052_at | F13A1 | 5.346 | 0.0027 | 11.69181717 | ‘Coagulation factor XIII, A1 polypeptide’ |
| 35220_at | ENPEP | 4.200 | 0.0027 | 11.66308093 | Glutamyl aminopeptidase (aminopeptidase A) |
| 37512_at | RODH | 5.112 | 0.0027 | 11.65031202 | 3-Hydroxysteroid epimerase |
| 103_at | THBS4 | 5.114 | 0.0027 | 11.6063075 | Thrombospondin 4 |
| 36156_at | AQP1 | 6.927 | 0.0027 | 11.6016663 | ‘Aquaporin 1’ |
| 39593_at | FGL2 | 5.176 | 0.0027 | 11.38223584 | Fibrinogen-like 2 |
| 33248_at | HOXA11 | 6.555 | 0.0027 | 11.34828098 | Homeo box A11 |
| 41420_at | IGFBP5 | 4.166 | 0.0041 | 11.32365037 | Insulin-like growth factor binding protein 5 |
| 35644_at | HEPH | 6.666 | 0.0027 | 11.26324425 | Hephaestin |
| 39646_at | CACNB2 | 4.719 | 0.0027 | 11.25017414 | ‘Calcium channel, voltage-dependent, beta 2 subunit’ |
| 35333_r_at | DVS27 | 4.403 | 0.0027 | 11.23759398 | DVS27-related protein |
| 33182_at | — | 4.990 | 0.0027 | 11.20258783 | Homo sapiens mRNA full-length insert cDNA clone EUROIMAGE 1630957 |
| 34303_at | FLJ90798 | 4.909 | 0.0027 | 11.10458088 | Hypothetical protein FLJ90798 |
| 33756_at | AOC3 | 5.186 | 0.0027 | 11.09776026 | ‘Amine oxidase, copper containing 3 (vascular adhesion protein 1)’ |
| 37710_at | MEF2C | 7.485 | 0.0027 | 11.03832245 | ‘MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C)’ |
| 35680_r_at | DPP6 | 3.783 | 0.0027 | 10.88151987 | Dipeptidylpeptidase 6 |
| 40126_at | — | 4.620 | 0.0027 | 10.87424764 | — |
| 31831_at | SMTN | 5.712 | 0.0027 | 10.84917408 | Smoothelin |
| 234_s_at | PTN | 5.660 | 0.0041 | 10.803161 | ‘Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1)’ |
| 36939_at | GPM6A | 4.981 | 0.0027 | 10.76644676 | Glycoprotein M6A |
| 41158_at | PLP1 | 4.879 | 0.0027 | 10.76462278 | ‘Proteolipid protein 1 (Pelizaeus-Merzbacher disease, spastic paraplegia 2, uncomplicated)’ |
| 41839_at | GAS1 | 4.401 | 0.0027 | 10.75375673 | Growth arrest-specific 1 |
| 1186_at | GDF10 | 4.165 | 0.0063 | 10.74677086 | Growth differentiation factor 10 |
| 35404_at | TACR2 | 4.416 | 0.0063 | 10.67366905 | Tachykinin receptor 2 |
| 160023_at | WNT2 | 5.424 | 0.0027 | 10.67077677 | Wingless-type MMTV integration site family member 2 |
| 34561_at | MS4A2 | 6.380 | 0.0027 | 10.6336763 | ‘Membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide)’ |
| 36280_at | GZMK | 4.739 | 0.0027 | 10.5607572 | ‘Granzyme K’ |
| 35668_at | RAMP1 | 3.430 | 0.0027 | 10.4886967 | Receptor (calcitonin) activity modifying protein 1 |
| 32521_at | SFRP1 | 4.488 | 0.0027 | 10.43051698 | Secreted frizzled-related protein 1 |
| 1975_s_at | IGF1 | 7.381 | 0.0027 | 10.34307322 | Insulin-like growth factor 1 (somatomedin C) |
| 37671_at | LAMA4 | 5.094 | 0.0027 | 10.34084543 | ‘Laminin, alpha 4’ |
| 40013_at | CLIC2 | 5.388 | 0.0027 | 10.2736905 | Chloride intracellular channel 2 |
| 32782_r_at | BPAG1 | 4.597 | 0.0027 | 10.2197061 | ‘Bullous pemphigoid antigen 1, 230/240 kDa’ |
| 36918_at | GUCY1A3 | 4.344 | 0.0027 | 10.18606373 | ‘Guanylate cyclase 1, soluble, alpha 3’ |
| 32239_at | MATN2 | 6.197 | 0.0027 | 10.14895891 | Matrilin 2 |
| 36503_at | CCL21 | 6.812 | 0.0027 | 10.09354903 | Chemokine (C–C motif) ligand 21 |
| 38508_s_at | TNXB | 8.826 | 0.0027 | 10.0794719 | Tenascin XB |
| 35146_at | TGFB1I1 | 5.562 | 0.0027 | 10.0680837 | Transforming growth factor beta 1 |
| 38653_at | PMP22 | 6.085 | 0.0027 | 10.04665889 | Peripheral myelin protein 22 |
| 342_at | ENPP1 | 5.008 | 0.0041 | 10.0434301 | Ectonucleotide pyrophosphatase/phosphodiesterase |
| 32826_at | ENTPD1 | 6.423 | 0.0027 | 10.01186286 | Ectonucleoside triphosphate diphosphohydrolase 1 |
| 40318_at | DNCI1 | 3.348 | 0.0063 | 10.00454416 | ‘Dynein, cytoplasmic, intermediate polypeptide 1’ |
Validation of the microarray data
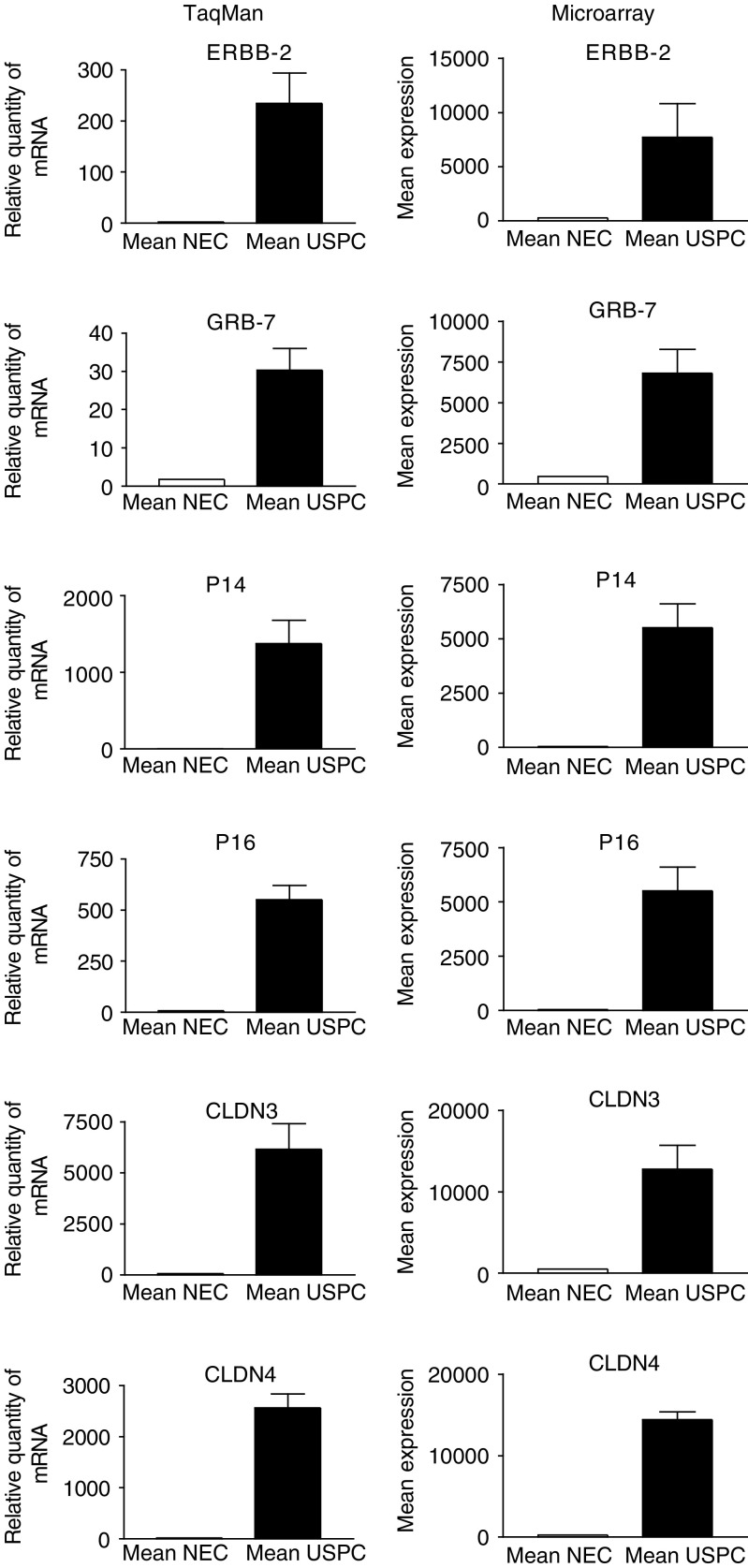
We used q-RT–PCR assays to validate the microarray data. Seven highly differentially expressed genes between USPC and NEC (i.e., CDKN2A/p16, CDKN2A/p14ARF (101-fold), L1CAM (25-fold), claudin-3 (eight-fold), claudin-4 (12-fold), GRB-7 (19-fold), and c-erbB2 (14-fold)) were selected for q-RT–PCR analysis. A comparison of the microarray and q-RT–PCR data for six of these genes is shown in Figure 2. Expression differences between USPC and NEC for CDKN2A/p16 (P=0.002), CDKN2A/p14ARF (P=0.002), claudin-3 (P=0.01), claudin-4 (P=0.002), GRB-7 (P=0.002) and c-erbB2 (P=0.01) were readily apparent (Table 2 and Figure 2). Moreover, for all seven genes tested, the q-RT–PCR data were highly correlated to the microarray data (P<0.001) (r=0.81, 0.80, 0.75, 0,69, 0.82, 0.71 and 0.65, respectively), with all the samples (i.e., 10 USPC and five NEC) included in both the q-RT–PCR and microarray experiments. Thus, q-RT–PCR data suggest that most array probe sets are likely to accurately measure the levels of the intended transcript within a complex mixture of transcripts.
Figure 2.
Quantitative RT–PCR and microarray expression analysis of CDKN2A/p16, CDKN2A/p14ARF, claudin-3, claudin-4, GRB-7 and c-erbB2 genes differentially expressed between USPC and NEC. Quantitative RT-PCR data were highly correlated to the microarray data (P<0.001).
Claudin-4 expression by immunohistology on USPC and NEC tissue blocks
To determine whether the high or low expression of the claudin-4 gene detected by microarray and q-RT–PCR assays in primary USPC and NEC cell lines, respectively, is the result of a selection of a subpopulation of cancer cells present in the original tumour, or whether in vitro expansion conditions may have modified gene expression, we performed immunohistochemical analysis of claudin-4 protein expression on formalin-fixed tumour tissue from the uncultured primary surgical specimens from which the USPC cell lines were derived. As shown in Table 4 and representatively in Figure 3, both cytoplasmic and membranous staining for claudin-4 protein expression was noted in the majority of USPC specimens (i.e., 90% score 3+ and 2+). In contrast, only low levels of membranous staining for claudin-4 protein was found in the NEC tissue samples tested by immunohistochemistry (Table 4, Figure 3, P=0.02 USPC vs NEC by Student t-test). To confirm and validate the immunohistochemical results in an independent series of USPC, formalin-fixed tumour tissue blocks from eight further surgical specimens (i.e., USPC 11–18, Table 4) similarly obtained from patients harbouring advanced stage disease were tested for claudin-4 expression. Again, heavy cytoplasmic and/or membranous staining for the claudin-4 receptor was found in the striking majority of the further USPC sample tested.
Table 4. Claudin-4 staining.
| Patient | Claudin-4 positivity |
|---|---|
| NEC 1 | 1+ |
| NEC 2 | 1+ |
| NEC 3 | 1+ |
| NEC 4 | 1+ |
| NEC 5 | 1+ |
| USPC 1 | 3+ |
| USPC 2 | 3+ |
| USPC 3 | 3+ |
| USPC 4 | 2+ |
| USPC 5 | 3+ |
| USPC 6 | 2+ |
| USPC 7 | 3+ |
| USPC 8 | 1+ |
| USPC 9 | 3+ |
| USPC 10 | 3+ |
| USPC 11 | 3+ |
| USPC 12 | 3+ |
| USPC 13 | 1+ |
| USPC 14 | 2+ |
| USPC 15 | 3+ |
| USPC 16 | 2+ |
| USPC 17 | 3+ |
| USPC 18 | 2+ |
Figure 3.
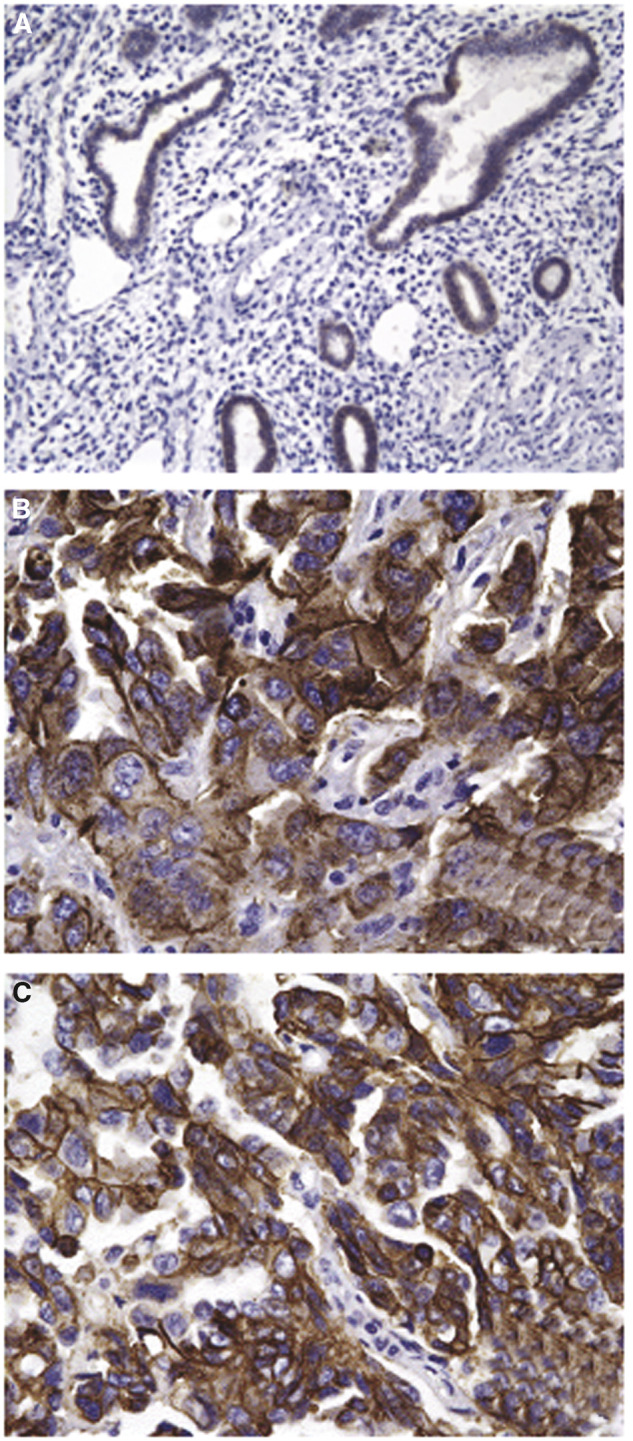
Representative immunohistochemical staining for claudin-4 on two paraffin-embedded USPC specimens (B–C) and one NEC specimen (A). Normal endometrial cell 1 (upper panel) showed light membrane staining for claudin-4, while USPC 1 and USPC 3 showed heavy cytoplasmic and membranous staining for claudin-4 (middle and lower panel). Note the large difference in the size of USPC cells when compared to the control glandular epithelium of a menopausal women. Original magnification × 400.
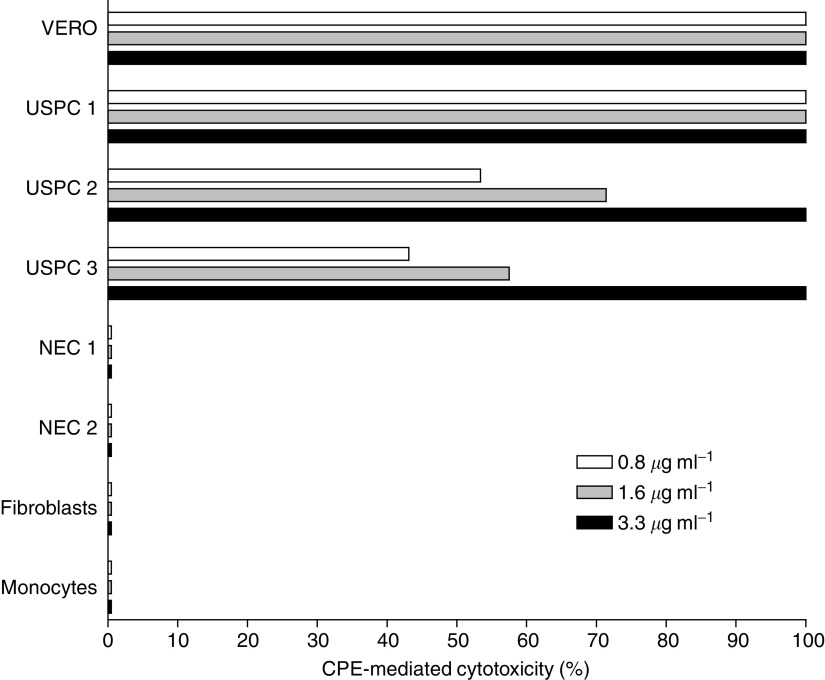
Effects of CPE on primary USPC and normal control cells
The sensitivity of primary uterine serous tumour cultures to CPE-mediated cytolysis was tested along with an appropriate claudin-3- and claudin-4-expressing positive control (i.e., Vero cells, obtained from the American Type Culture Collection), and negative controls which do not express detectable levels of either claudin-3 or claudin-4. As shown in Figure 4, all primary USPC tested were found highly sensitive to CPE-mediated cytolysis. The cytotoxic effect was dose dependent and was positively correlated to the levels of either claudin-3 or claudin-4 expression as tested by RT–PCR in tumor samples. Importantly, although uterine serous tumours demonstrated different sensitivities to CPE exposure, no USPC was found viable after 24 h exposure to CPE at the concentration of 3.3 μg ml−1. In contrast, all normal controls tested, including endometrial epithelium, fibroblasts and mononuclear cells lacking claudin-3 or claudin-4, were not affected by CPE (Figure 4).
Figure 4.
Representative dose-dependent CPE-mediated cytotoxicity of primary USPC compared to positive control Vero cells or negative controls (i.e., NEC, fibroblasts, and monocytes) after 24 h exposure to scalar doses of CPE. VERO, positive control cells. Uterine serous papillary carcinoma-1 to USPC-3 primary uterine serous tumors. NEC, normal endometrial cells. Fibroblasts, normal human fibroblasts. Monocytes, normal mononuclear cells.
DISCUSSION
This report represents the first communication of an investigation involving the genome-wide examination of differences in gene expression between primary USPC and normal endometrial cells (NEC). In this study, we have used short-term primary USPC and NEC cultures (to minimise the risk of a selection bias inherent in any long-term in vitro growth) to study differential gene expression in highly enriched populations of epithelial tumour cells and normal cells. We found that hierarchical clustering of the samples and gene expression levels within the samples led to the unambiguous separation of USPC from NEC. We detected 529 genes differentially expressed between USPC and NEC, whose average change in expression level between the two groups was at least five-fold and which were found significant with both WRS test and SAM analysis. The known function of some of these genes may provide insights into the molecular pathogenesis and the highly aggressive biologic behaviour of uterine serous tumours, while others may prove to be useful diagnostic and therapeutic markers against this disease.
For example, the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene was found to be the most highly differentially expressed gene in USPC with over 101-fold upregulation relative to NEC. Importantly, the CDKN2A gene is a putative oncosuppressor gene encoding two unrelated proteins, both cellular growth inhibitors, in different reading frames (Quelle et al, 1995). One is p16, which regulates retinoblastoma protein (pRb)-dependent G1 arrest, and the second is p14ARF, which blocks MDM2-induced p53 degradation, resulting in an increase in p53 levels and consequent cell cycle arrest (Quelle et al, 1995). Although loss of p53 function is considered critical for the molecular pathogenesis of USPC (Moll et al, 1996), it is only recently that abnormality of the Rb pathway has been suggested to define a subgroup of aggressive endometrial carcinomas with poor prognosis (Salvesen et al, 2000). Quantitative RT-PCR analysis of expression of both p16 and p14ARF in our USPC series found extremely high levels of both transcripts, suggesting that the marked overexpression of the CDKN2A gene may be attributable to a negative feedback loop due to the loss of function of both pRb and p53 proteins (Moll et al, 1996; Salvesen et al, 2000). Consistent with this view, an inverse relationship between the expression of p16 and p14ARF proteins and the presence of normal or functional Rb and p53 in human cancer cells has been previously demonstrated (Khleif et al, 1996). Thus, our data suggest for the first time that CDKN2A gene overexpression may represent a consistent genetic anomaly of USPC secondary to an autoregulatory feedback loop due to disruption of both the p16-CDK4/cyclin D1-pRb pathway and the p14ARF-MDM2-p53 pathway.
Among the several potential therapeutic target gene products identified, genes encoding tight junction (TJ) proteins claudin-3 and claudin-4 were consistently found as two of the most highly upregulated genes in USPC, with over eight- and 12-fold upregulation, respectively, relative to NEC. To the best of our knowledge, claudin-3 and claudin-4 overexpression has not been previously linked to USPC. Although the exact function of claudin-3 and claudin-4 in USPC is still unclear, these proteins have recently been shown to represent the epithelial receptors for CPE (Katahira et al, 1997), and to be the only family members of the transmembrane tissue-specific claudin proteins capable of mediating CPE binding and cytolysis. As CPE may trigger a multistep mechanism leading to efficient lysis of mammalian target cells overexpressing claudin-3 and claudin-4 by an increase in membrane permeability resulting in loss of osmotic equilibrium (McClane 1996), CPE-mediated therapy might be a novel, potentially highly effective, strategy for the treatment of USPC refractory to chemotherapy as well as other human tumours overexpressing claudin-3 and/or claudin-4 (Long et al, 2001; Michl et al, 2001). Mammalian cells that do not express either claudin-3 and/or claudin-4 fail to bind CPE and are not susceptible to CPE cytotoxicity (McClane 1996). Consistent with this view, all primary USPC evaluated, including those found to be resistant to chemotherapy in vivo (i.e., USPC-1 and USPC-2, data not shown), were found highly sensitive to CPE-mediated killing in vitro. This was in strong contrast with the lack of sensitivity of normal control cells to CPE-mediated cytolysis. Although USPC tumours demonstrated different sensitivity to CPE exposure, no cancer was found viable after 24 h exposure to CPE at a concentration of 3.3 μg ml−1, a dose well tolerated by in vivo administration of CPE in murine animal models (Wallace et al, 1999). More extensive studies will be necessary to evaluate the potential and feasibility of CPE therapy in vivo in human patients. Nevertheless, our results provide strong evidence to suggest that CPE-based therapy may have great potential in the treatment of USPC patients refractory to standard treatment modalities. Protein expression data obtained by immunohistochemistry with anti-claudin-4 antibody on an independent set of USPC blocks further support the proposal that claudins may represent therapeutic targets.
The organization of kallikreins, a gene family now consisting of 15 genes that encode for trypsin-like or chymotrypsin-like serine proteases, has been recently elucidated (Diamandis and Yousef, 2002). Serine proteases have well-characterized roles in diverse cellular activities, including blood coagulation, wound healing, digestion, and immune responses, as well as tumour invasion and metastasis (reviewed in Diamandis and Yousef, 2002). Secreted serine proteases such as prostate-specific antigen (PSA) and kallikrein 2 have already found important clinical application as prostate cancer biomarkers (Diamandis and Yousef, 2002). Of interest, kallikrein 6 (also known as zyme/protease M/neurosin) and kallikrein-10 (NES1), two serine proteases recently shown to be present at high levels in the circulation of a subset of ovarian cancer patients (Diamandis et al, 2003; Luo et al, 2003), were both highly differentially expressed genes in USPC when compared to NEC. Kallikrein 6 and kallikrein 10 overexpression has been shown to correlate with intrinsic resistance to adjuvant chemotherapy and with a poor prognosis in ovarian cancer patients (Diamandis et al, 2003; Luo et al, 2003). These data are thus consistent with our results showing high expression of kallikrein 6 and kallikrein 10 in USPC, a variant of endometrial carcinoma characterised by an aggressive biologic behaviour and an inborn resistance to chemotherapy (Nicklin and Copeland, 1996). In addition, these results further emphasise the view that kallikrein 6 and kallikrein 10 have the potential to become novel cancer markers for early diagnosis and/or monitoring of USPC, as well as possible immunotherapeutic targets of vaccination strategies against recurrent/refractory serous papillary gynecologic tumours (Cannon et al, 2002).
c-erbB2 gene was found to be one of the most highly differentially expressed gene in USPC with over 14-fold upregulation compared with NEC. Furthermore, the growth factor receptor-bound protein 7 (GRB7), a gene tightly linked to c-erbB2 and previously reported coamplified and coexpressed with this gene in several cancer types (Janes et al, 1997), was also highly differentially expressed in USPC compared to NEC. These data confirm our recent discovery of a striking overexpression of the c-erbB2 gene product HER2/neu on 80% of pure USPC (Santin et al, 2002). Thus, HER2/neu overexpression may represent a distinctive molecular marker that, in addition to having the potential to facilitate differentiation of USPC from the histologically indistinguishable high-grade serious ovarian tumours (Santin et al, 2004), may also provide insights into the disproportionately poor prognosis of USPC patients (Lukes et al, 1994; Santin et al, 2002, 2004). Previous studies have reported that HER2/neu overexpression in USPC patients may be associated with resistance to chemotherapeutic drugs and shorter survival (Lukes et al, 1994). However, high overexpression of the c-erbB2 gene on USPC provides support for the notion that trastuzumab (Herceptin, Genentech, San Francisco, CA, USA), a humanised anti-HER-2/Neu antibody that is showing great promise for treatment of metastatic breast cancer patients overexpressing HER-2/Neu protein (Slamon et al, 2001), may be a novel, potentially highly effective therapy against USPC. Consistent with this view, high sensitivity of USPC to natural killer (NK) cell-mediated antibody-dependent cytotoxicity triggered by anti-HER-2/Neu-specific antibody in vitro (Santin et al, 2002), as well as clinical responses in vivo (Villella et al, 2003), have recently been reported with the use of Herceptin in USPC patients.
Several other highly ranked genes have been identified in our USPC gene expression profiling analysis, including membrane-associated protein 17 (MAP17), galanin, urokinase plasminogen activator receptor (UPAR), interleukin-6, forkhead box M1, interleukin-18, dickkopf homolog 1 (DKK1), coagulation factor II (thrombin) receptor-like 1, transforming growth factor, alpha, interleukin 8, topoisomerase (DNA) II alpha, hyaluronan-mediated motility receptor (RHAMM), and secretory leukocyte protease inhibitor (antileukoproteinase). For most of the genes found differentially expressed in our experiments, a correlation with USPC cancer development and progression has not been recognised before. DKK1, for example, has been recently reported by our group to play an important role in the development of osteolytic lesions in multiple myeloma (Tian et al, 2003), but its possible role in USPC pathogenesis and/or progression has not been elucidated. For other genes such as UPAR, a glycosyl-phosphatidylinositol-anchored glycoprotein whose role in promoting tumour cell invasion and metastases has been well established in a number of experimental studies, a correlation with high expression in the USPC phenotype has been recently reported (Memarzadeh et al, 2002).
A large number of downregulated (at least five-fold) genes in USPC vs NEC, such as transforming growth factor beta receptor III, platelet-derived growth factor receptor alpha, SEMACAP3, ras homolog gene family member I (ARHI), and DOC1 (Table 3), have been identified in our analysis. Some of these genes encode for widely held tumor suppressor genes such as SEMACAP3, ARHI, and DOC1 (Liu and Ganesan, 2002), others for proteins important for tissue homeostasis or that have been previously implicated in apoptosis, proliferation, adhesion, or tissue maintenance. Owing to space limitations, we will not comment further upon the cluster of genes that showed downregulation of the transcripts in invasive tumours.
In conclusion, multiple USPC restricted markers have been identified through our analysis. Most of these genes have not been previously linked with this disease and thus represent novel findings. The identification of HER2/neu and CPE epithelial receptors among others as some of the most highly differentially expressed genes in USPC when compared to NEC suggest that therapeutic strategies targeting HER2/neu by monoclonal antibodies (Villella et al, 2003) and claudin-3 and claudin-4 by local and/or systemic administration of CPE (Long et al, 2001; Michl et al, 2001) may represent novel potentially effective modalities for the treatment of patients harbouring this highly aggressive and chemotherapy-resistant variant of endometrial cancer. The future design and implementation of clinical trials at this regard will ultimately determine the validity of these approaches.
References
- Bohkman JV (1983) Two pathogenetic types of endometrial carcinoma. Gynecol Oncol 15: 10–17 [DOI] [PubMed] [Google Scholar]
- Bongso A, Gajra B, Lian NP, Wong PC, Soon-Chye N, Ratnam S (1988) Establishment of human endometrial cell cultures. Human Reprod 3: 705–713 [DOI] [PubMed] [Google Scholar]
- Cannon MJ, O'Brien TJ, Underwood LJ, Crew MD, Bondurant KL, Santin AD (2002) Novel target antigens for dendritic cell-based immunotherapy against ovarian cancer. Expert Rev Anticancer Ther 2: 97–105 [DOI] [PubMed] [Google Scholar]
- Diamandis EP, Scorilas A, Fracchioli S, Van Gramberen M, De Bruijn H, Henrik A, Soosaipillai A, Grass L, Yousef GM, Stenman UH, Massobrio M, Van Der Zee AG, Vergote I, Katsaros D (2003) Human kallikrein 6 (hK6): a new potential serum biomarker for diagnosis and prognosis of ovarian carcinoma. J Clin Oncol 21: 1035–1043 [DOI] [PubMed] [Google Scholar]
- Diamandis EP, Yousef GM (2002) Human tissue kallikreins: a family of new cancer biomarkers. Clin Chem 48: 1198–1205 [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson M, Ross J, Eifel P, Martinez A, Kempson R 1982. Uterine papillary serous carcinoma: a highly malignant form of endometrial adenocarcinoma. Am J Surg Pathol 6: 93–108 [DOI] [PubMed] [Google Scholar]
- Janes PW, Lackmann M, Church WB, Sanderson GM, Sutherland RL, Daly RJ (1997) Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. J Biol Chem 272: 8490–8497 [DOI] [PubMed] [Google Scholar]
- Jemal A, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ 2003. Cancer statistics. CA Cancer J Clin 53: 5–26 [DOI] [PubMed] [Google Scholar]
- Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N (1997) Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem 272: 26652–26658 [DOI] [PubMed] [Google Scholar]
- Khleif SN, DeGregori J, Yee CL, Otterson GA, Kaye FJ, Nevins JR, Howley PM (1996) Inhibition of cyclin D-CDK4/CDK6 activity is associated with an E2F-mediated induction of cyclin kinase inhibitor activity. Proc Natl Acad Sci USA 93: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ganesan TS (2002) Tumour suppressor genes in sporadic epithelial ovarian cancer. Reproduction 123: 341–353 [DOI] [PubMed] [Google Scholar]
- Long H, Crean CD, Lee WH, Cummings OW, Gabig TG (2001) Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res 61: 7878–7881 [PubMed] [Google Scholar]
- Lukes AS, Kohler MF, Pieper CF, Kerns BJ, Bentley R, Rodriguez GC, Soper JT, Clarke-Pearson DL, Bast Jr RC, Berchuck A (1994) Multivariable analysis of DNA ploidy, p53, and HER-2/neu as prognostic factors in endometrial cancer. Cancer 73: 2380–2385 [DOI] [PubMed] [Google Scholar]
- Luo LY, Katsaros D, Scorilas A, Fracchioli S, Bellino R, van Gramberen M, de Bruijn H, Henrik A, Stenman UH, Massobrio M, van der Zee AG, Vergote I, Diamandis EP (2003) The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res 63: 807–811 [PubMed] [Google Scholar]
- Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minaguchi T, Kuramoto H, Nishida M, Satoh T, Tanaka T, Nakamura Y (2001) Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN. Cancer Res 61: 3741–3749 [PubMed] [Google Scholar]
- McClane BA (1996) An overview of Clostridium perfringens enterotoxin. Toxicon 34: 1335–1343 [DOI] [PubMed] [Google Scholar]
- Memarzadeh S, Kozak KR, Chang L, Natarajan S, Shintaku P, Reddy ST, Farias-Eisner R, Memarzedeh S (2002) Urokinase plasminogen activator receptor: prognostic biomarker for endometrial cancer. Proc Natl Acad Sci USA 99: 10647–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM (2001) Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 121: 678–684 [DOI] [PubMed] [Google Scholar]
- Moll UM, Chalas E, Auguste M, Meaney D, Chumas J (1996) Uterine papillary serous carcinoma evolves via a p53-driven pathway. Hum Pathol 27: 1295–1300 [DOI] [PubMed] [Google Scholar]
- Moreno-Bueno G, Sanchez-Estevez C, Cassia R, Rodriguez-Perales S, Diaz-Uriarte R, Dominguez O, Hardisson D, Andujar M, Prat J, Matias-Guiu X, Cigudosa JC, Palacios J (2003) Differential gene expression profile in endometrioid and nonendometrioid endometrial carcinoma: STK15 is frequently overexpressed and amplified in nonendometrioid carcinomas. Cancer Res 63: 5697–5702 [PubMed] [Google Scholar]
- Nicklin JL, Copeland LJ (1996) Endometrial papillary serous carcinoma: pattern of spread and treatment. Clin Obstetr Gynecol 39: 686–695 [DOI] [PubMed] [Google Scholar]
- Quelle DE, Zindy F, Ashmun RA, Sherr CJ (1995) Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83: 993–1000 [DOI] [PubMed] [Google Scholar]
- Risinger JI, Maxwell GL, Chandramouli GV, Jazaeri A, Aprelikova O, Patterson T, Berchuck A, Barrett JC (2003) Microarray analysis reveals distinct gene expression profiles among different histologic types of endometrial cancer. Cancer Res 63: 6–11 [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H (2002) Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 346: 1937–1947 [DOI] [PubMed] [Google Scholar]
- Salvesen HB, Das S, Akslen LA. (2000) Loss of nuclear p16 protein expression is not associated with promoter methylation but defines a subgroup of aggressive endometrial carcinomas with poor prognosis. Clin Cancer Res 6: 153–159 [PubMed] [Google Scholar]
- Santin AD, Bellone S, Gokden M, Palmieri M, Dunn D, Agha J, Roman JJ, Hutchins L, Pecorelli S, O'Brien T, Cannon MJ, Parham GP (2002) Overexpression of HER-2/neu in uterine serous papillary cancer. Clin Cancer Res 8: 1271–1279 [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane' S, Gokden M, Roman JJ, O'Brien T, Tian E, Cannon MJ, Shaughnessy J, Pecorelli S (2004) Discrimination between uterine serous papillary carcinomas and ovarian serous papillary tumors by gene expression profiling. Br J Cancer 90: 1814–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamon DL, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L (2001) Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344: 783–789 [DOI] [PubMed] [Google Scholar]
- Smid-Koopman E, Blok LJ, Chadha-Ajwani S, Helmerhorst TJ, Brinkmann AO, Huikeshoven FJ (2000) Gene expression profiles of human endometrial cancer samples using a cDNA-expression array technique: assessment of an analysis method. Br J Cancer 83: 246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Van de Rijn M, Jeffrey SS, Thorsen T, Quist H (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy Jr JD (2003) The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. New Engl J Med 349: 2483–2494 [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella JA, Cohen S, Tiersten A, Smith DH, Hibshoosh H, Hershman D (2003) Her2/neu expression in uterine papillary serous cancers. Proc Am Soc Clin Oncol 22: 465 [Google Scholar]
- Wallace FM, Mach AS, Keller AM, Lindsay JA (1999) Evidence for Clostridium perfringens enterotoxin (CPE) inducing a mitogenic and cytokine response in vitro and a cytokine response in vivo. Curr Microbiol 38: 96–100 [DOI] [PubMed] [Google Scholar]
- Zhan F, Tian E, Bumm K, Smith R, Barlogie B, Shaughnessy Jr J (2002) Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood 99: 1745–1757 [DOI] [PubMed] [Google Scholar]