Abstract
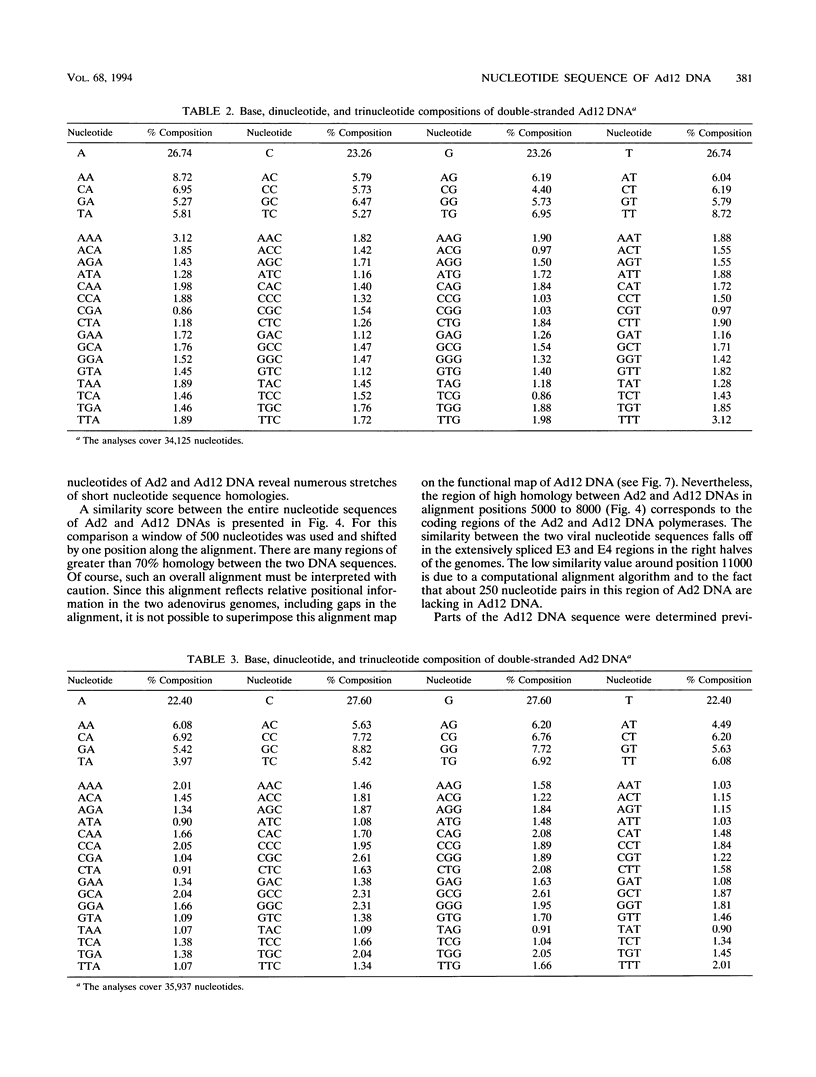
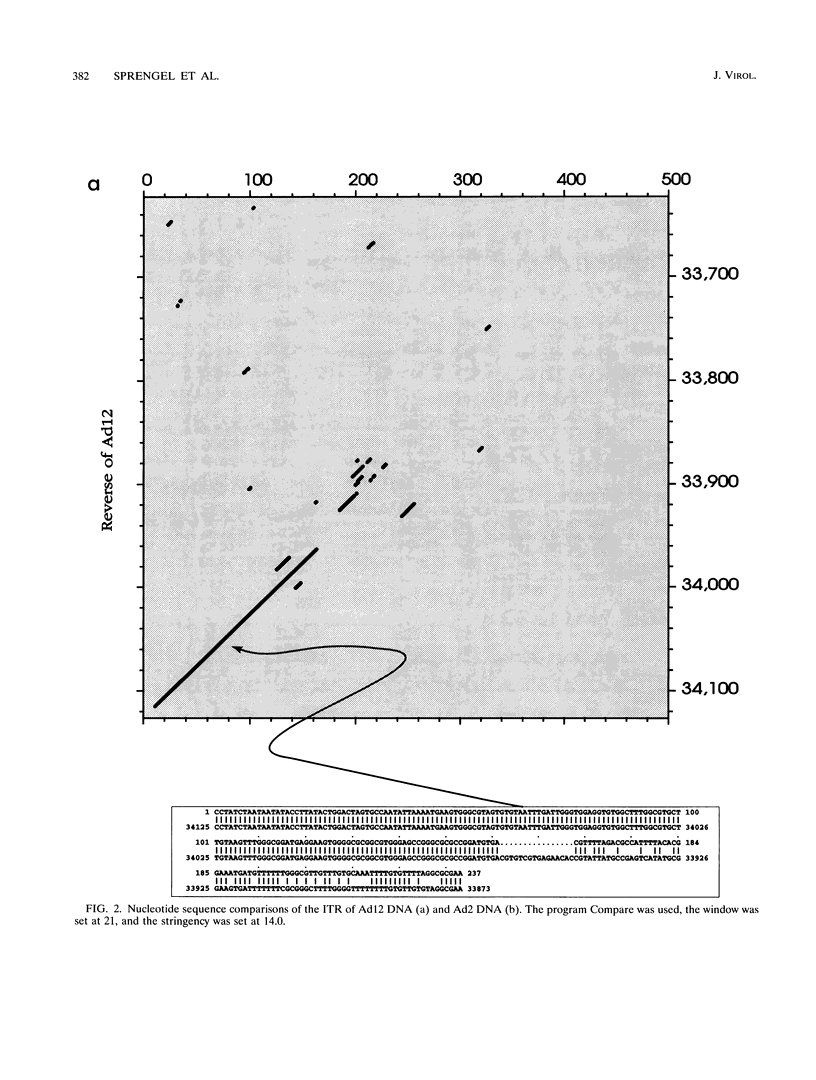
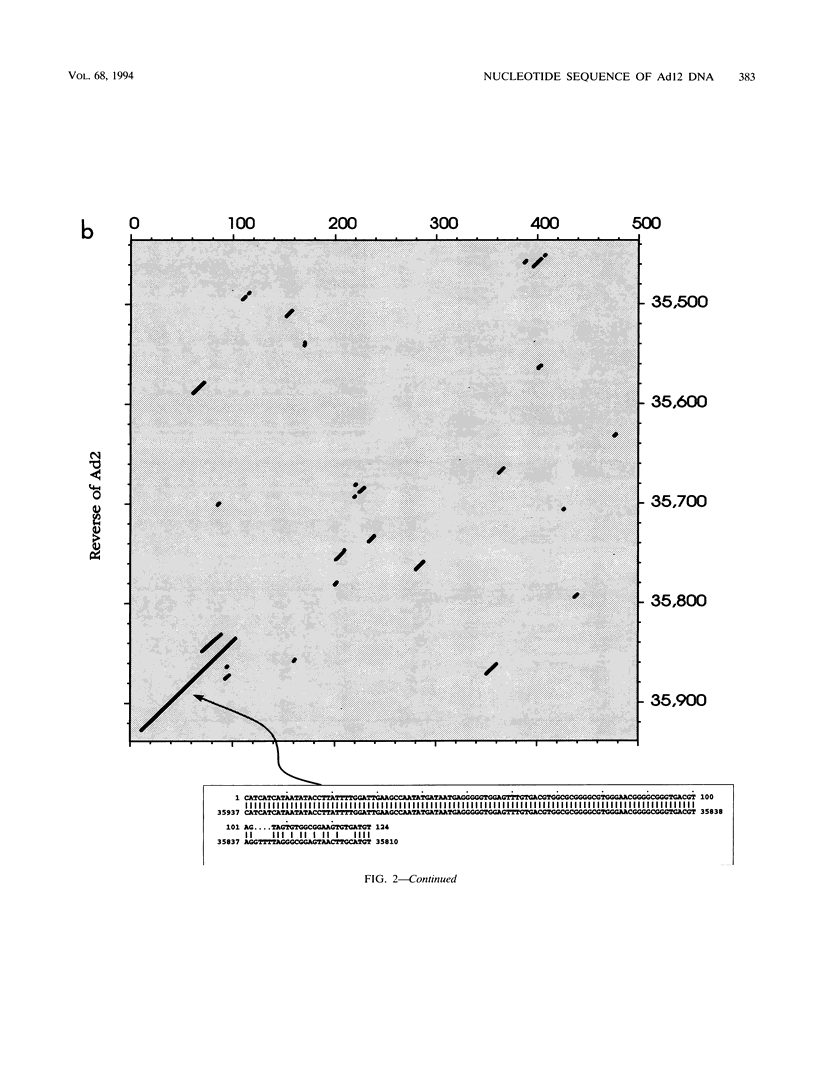
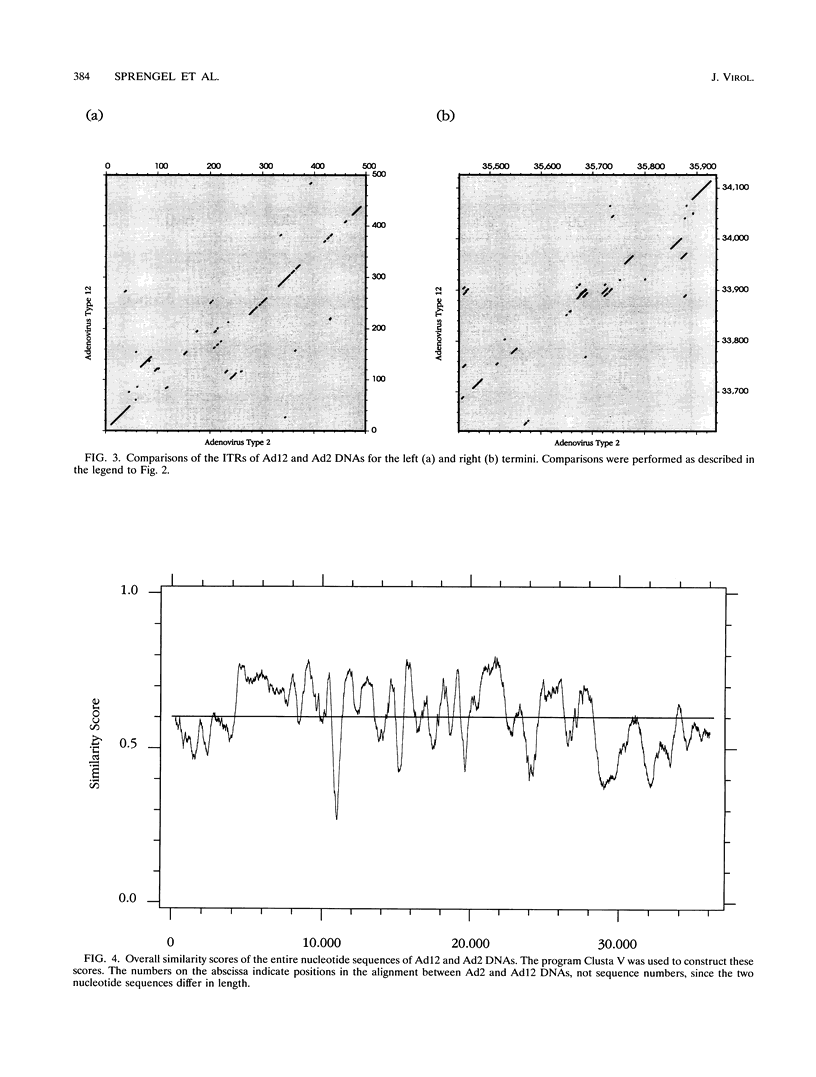
A fresh inoculum of human adenovirus type 12 (Ad12) was obtained from the American Type Culture Collection and passaged once on human embryonic kidney cells, and Ad12 DNA was prepared from the first-passage yield to avoid higher passages which might have generated host-virus DNA recombinants. The 18 PstI fragments of Ad12 DNA were cloned into the pBluescript KS vector, and the entire nucleotide sequence of both strands from all 18 fragments was determined by using successive oligodeoxyribonucleotide primers. Ad12 DNA extends over 34,125 nucleotide pairs, and its molecular weight is calculated to be about 22 x 10(6). The nucleotide sequence of Ad12 DNA was subjected to computer analyses that determined possible open reading of frames on the two strands, the leader sequences, the position of the virus-associated RNA coding region, possible TATA, and polyadenylation signals. The distribution of the Ad12 open reading frames was similar to that in the previously sequenced Ad2 DNA, but there were also distinct differences. Ad12 DNA has an inverted terminal redundancy of 161 nucleotides, compared with 102 nucleotides in Ad2 DNA. There were stretches of sequence identity between Ad2 and Ad12 DNAs at both termini; the overall sequence similarity between the two viral genomes ranged between 59% (polypeptide IX) and 77% (in the E2 region), with high homology also in the sequences for the adenovirus DNA polymerase.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1992 May 11;20 (Suppl):2019–2022. doi: 10.1093/nar/20.suppl.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J. L., Polder L. J., Bernards R., Schrier P. I., van den Elsen P. J., van der Eb A. J., van Ormondt H. The 2.2 kb E1b mRNA of human Ad12 and Ad5 codes for two tumor antigens starting at different AUG triplets. Cell. 1981 Nov;27(1 Pt 2):121–131. doi: 10.1016/0092-8674(81)90366-4. [DOI] [PubMed] [Google Scholar]
- Bucher P., Trifonov E. N. Compilation and analysis of eukaryotic POL II promoter sequences. Nucleic Acids Res. 1986 Dec 22;14(24):10009–10026. doi: 10.1093/nar/14.24.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chroboczek J., Bieber F., Jacrot B. The sequence of the genome of adenovirus type 5 and its comparison with the genome of adenovirus type 2. Virology. 1992 Jan;186(1):280–285. doi: 10.1016/0042-6822(92)90082-z. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R., Doerfler W. Proof of recombination between viral and cellular genomes in human KB cells productively infected by adenovirus type 12: structure of the junction site in a symmetric recombinant (SYREC). Gene. 1983 Dec;26(2-3):283–289. doi: 10.1016/0378-1119(83)90198-1. [DOI] [PubMed] [Google Scholar]
- Deuring R., Klotz G., Doerfler W. An unusual symmetric recombinant between adenovirus type 12 DNA and human cell DNA. Proc Natl Acad Sci U S A. 1981 May;78(5):3142–3146. doi: 10.1073/pnas.78.5.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. Abortive infection and malignant transformation by adenoviruses: integration of viral DNA and control of viral gene expression by specific patterns of DNA methylation. Adv Virus Res. 1991;39:89–128. doi: 10.1016/s0065-3527(08)60793-9. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Gahlmann R., Stabel S., Deuring R., Lichtenberg U., Schulz M., Eick D., Leisten R. On the mechanism of recombination between adenoviral and cellular DNAs: the structure of junction sites. Curr Top Microbiol Immunol. 1984;109:193–228. doi: 10.1007/978-3-642-69460-8_9. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Hellmann W., Kleinschmidt A. K. The DNA of adenovirus type 12 and its denaturation pattern. Virology. 1972 Feb;47(2):507–512. doi: 10.1016/0042-6822(72)90290-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Nonproductive infection of baby hamster kidney cells (BHK21) with adenovirus type 12. Virology. 1969 Aug;38(4):587–606. doi: 10.1016/0042-6822(69)90179-2. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Patterns of DNA methylation--evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol Chem Hoppe Seyler. 1991 Aug;372(8):557–564. [PubMed] [Google Scholar]
- Doerfler W. The fate of the DNA of adenovirus type 12 in baby hamster kidney cells. Proc Natl Acad Sci U S A. 1968 Jun;60(2):636–643. doi: 10.1073/pnas.60.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esche H., Reuther M., Schughart K. Early and late proteins of adenovirus type 12: translation mapping with RNA isolated from infected and transformed cells. Curr Top Microbiol Immunol. 1984;111:91–106. doi: 10.1007/978-3-642-69549-0_4. [DOI] [PubMed] [Google Scholar]
- Esche H., Schilling R., Doerfler W. In vitro translation of adenovirus type 12-specific mRNA isolated from infected and transformed cells. J Virol. 1979 Apr;30(1):21–31. doi: 10.1128/jvi.30.1.21-31.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimuth P., Anderson C. W. Human adenovirus serotype 12 virion precursors pMu and pVI are cleaved at amino-terminal and carboxy-terminal sites that conform to the adenovirus 2 endoproteinase cleavage consensus sequence. Virology. 1993 Mar;193(1):348–355. doi: 10.1006/viro.1993.1131. [DOI] [PubMed] [Google Scholar]
- Ghosh D. TFD: the transcription factors database. Nucleic Acids Res. 1992 May 11;20 (Suppl):2091–2093. doi: 10.1093/nar/20.suppl.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hogenkamp T., Esche H. Nucleotide sequence of the right 10% of adenovirus type 12 DNA encoding the entire region E4. Nucleic Acids Res. 1990 May 25;18(10):3065–3066. doi: 10.1093/nar/18.10.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde A., Weber J. M. The primary structure of human adenovirus type 12 protease. Nucleic Acids Res. 1988 Jul 25;16(14B):7195–7195. doi: 10.1093/nar/16.14.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Sawada Y., Shinawawa M., Shimizu Y., Shiroki K., Shimojo H., Sugisaki H., Takanami M., Uemizu Y., Fujinaga K. Nucleotide sequence of the transforming early region E1b of adenovirus type 12 DNA: structure and gene organization, and comparison with those of adenovirus type 5 DNA. Nucleic Acids Res. 1981 Dec 11;9(23):6571–6589. doi: 10.1093/nar/9.23.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., van Schaik F. M., Speijer J. G., Sussenbach J. S. Structure and function of adenovirus DNA binding protein: comparison of the amino acid sequences of the Ad5 and Ad12 proteins derived from the nucleotide sequence of the corresponding genes. Virology. 1983 Jul 15;128(1):140–153. doi: 10.1016/0042-6822(83)90325-2. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Edgell M. H., Hutchison C. A., 3rd The nucleotide sequence recognized by the BstEII restriction endonuclease. Gene. 1980 Dec;12(1-2):171–174. doi: 10.1016/0378-1119(80)90029-3. [DOI] [PubMed] [Google Scholar]
- Orend G., Kuhlmann I., Doerfler W. Spreading of DNA methylation across integrated foreign (adenovirus type 12) genomes in mammalian cells. J Virol. 1991 Aug;65(8):4301–4308. doi: 10.1128/jvi.65.8.4301-4308.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortin J., Scheidtmann K. H., Greenberg R., Westphal M., Doerfler W. Transcription of the genome of adenovirus type 12. III. Maps of stable RNA from productively infected human cells and abortively infected and transformed hamster cells. J Virol. 1976 Nov;20(2):355–372. doi: 10.1128/jvi.20.2.355-372.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E., Reinke C., Yamamoto N., zur Hausen H. Terminal rearrangements in the genome of adenovirus type 12 mutants adapted to growth in two human tumor cell lines. Virology. 1982 Jan 15;116(1):284–296. doi: 10.1016/0042-6822(82)90420-2. [DOI] [PubMed] [Google Scholar]
- Shibata H., Zheng J. H., Koikeda S., Masamune Y., Nakanishi Y. Cis- and trans-acting factors for transcription of the adenovirus 12 E1A gene. Biochim Biophys Acta. 1989 Mar 1;1007(2):184–191. doi: 10.1016/0167-4781(89)90037-7. [DOI] [PubMed] [Google Scholar]
- Shinagawa M., Padmanabhan R. Comparative sequence analysis of the inverted terminal repetitions from different adenoviruses. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3831–3835. doi: 10.1073/pnas.77.7.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L. M., Hong J. S., Wei Y. F., Engler J. A. Nucleotide sequence of the genes encoded in early region 2b of human adenovirus type 12. Gene. 1986;46(2-3):187–195. doi: 10.1016/0378-1119(86)90403-8. [DOI] [PubMed] [Google Scholar]
- Stoehr P. J., Cameron G. N. The EMBL data library. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2227–2230. doi: 10.1093/nar/19.suppl.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugisaki H., Sugimoto K., Takanami M., Shiroki K., Saito I., Shimojo H., Sawada Y., Uemizu Y., Uesugi S., Fujinaga K. Structure and gene organization in the transformed Hind III-G fragment of Ad12. Cell. 1980 Jul;20(3):777–786. doi: 10.1016/0092-8674(80)90324-4. [DOI] [PubMed] [Google Scholar]
- Sutter D., Westphal M., Doerfler W. Patterns of integration of viral DNA sequences in the genomes of adenovirus type 12-transformed hamster cells. Cell. 1978 Jul;14(3):569–585. doi: 10.1016/0092-8674(78)90243-x. [DOI] [PubMed] [Google Scholar]
- Tatzelt J., Fechteler K., Langenbach P., Doerfler W. Fractionated nuclear extracts from hamster cells catalyze cell-free recombination at selective sequences between adenovirus DNA and a hamster preinsertion site. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7356–7360. doi: 10.1073/pnas.90.15.7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatzelt J., Scholz B., Fechteler K., Jessberger R., Doerfler W. Recombination between adenovirus type 12 DNA and a hamster preinsertion sequence in a cell-free system. Patch homologies and fractionation of nuclear extracts. J Mol Biol. 1992 Jul 5;226(1):117–126. doi: 10.1016/0022-2836(92)90128-7. [DOI] [PubMed] [Google Scholar]
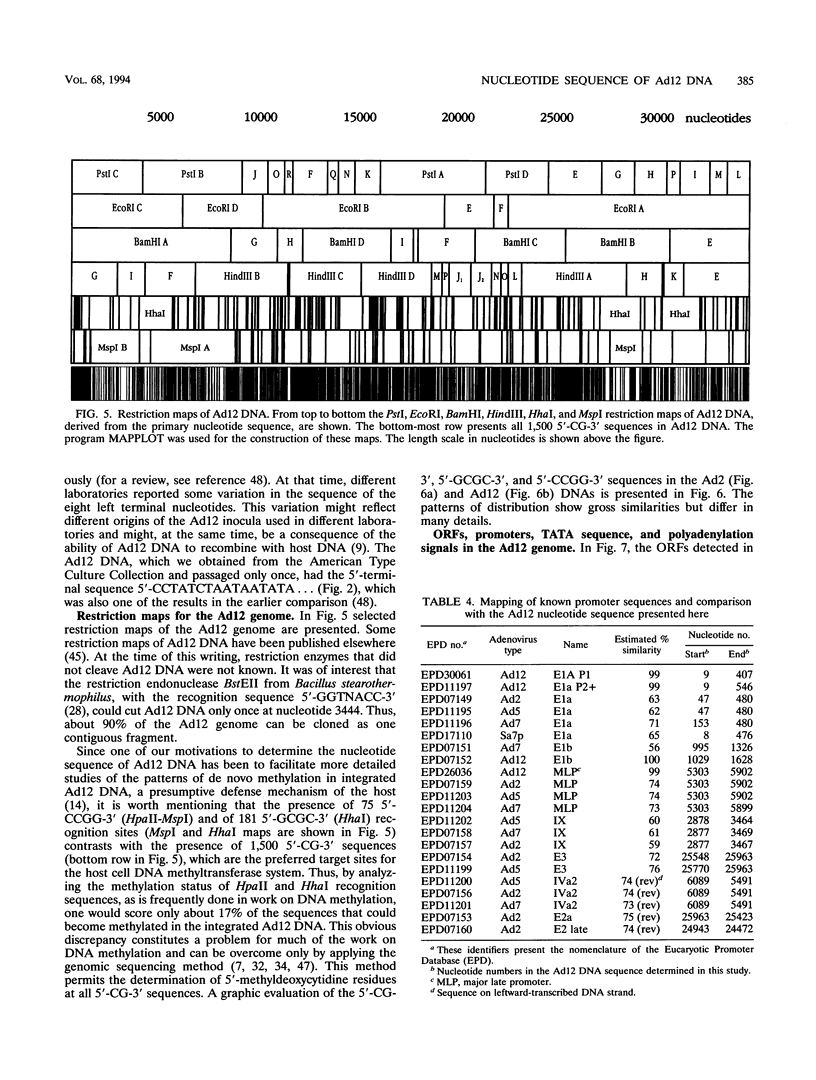
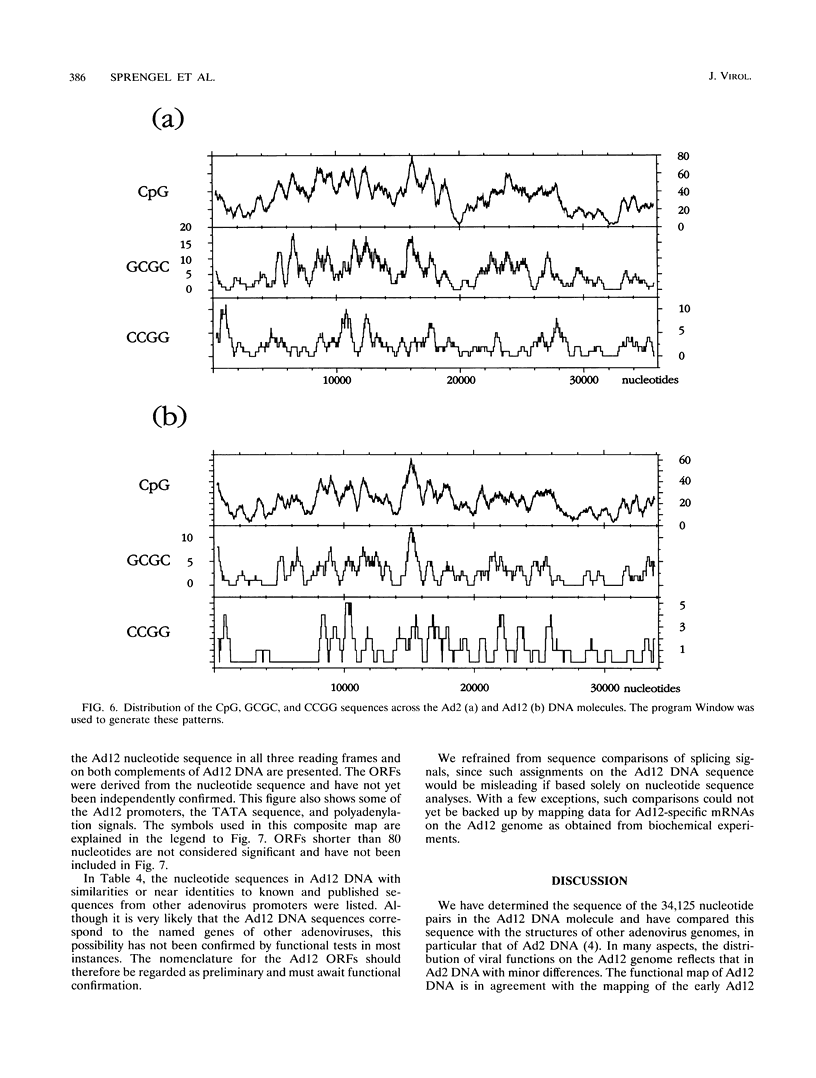
- Thörner A., Johansson M. E., Hierholzer J. C. Restriction endonuclease patterns of adenovirus type 12 and 18. J Virol Methods. 1992 Sep;39(1-2):101–109. doi: 10.1016/0166-0934(92)90129-2. [DOI] [PubMed] [Google Scholar]
- Tolun A., Aleström P., Pettersson U. Sequence of inverted terminal repetitions from different adenoviruses: demonstration of conserved sequences and homology between SA7 termini and SV40 DNA. Cell. 1979 Jul;17(3):705–713. doi: 10.1016/0092-8674(79)90277-0. [DOI] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci U S A. 1989 May;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zock C., Doerfler W. A mitigator sequence in the downstream region of the major late promoter of adenovirus type 12 DNA. EMBO J. 1990 May;9(5):1615–1623. doi: 10.1002/j.1460-2075.1990.tb08281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zock C., Iselt A., Doerfler W. A unique mitigator sequence determines the species specificity of the major late promoter in adenovirus type 12 DNA. J Virol. 1993 Feb;67(2):682–693. doi: 10.1128/jvi.67.2.682-693.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
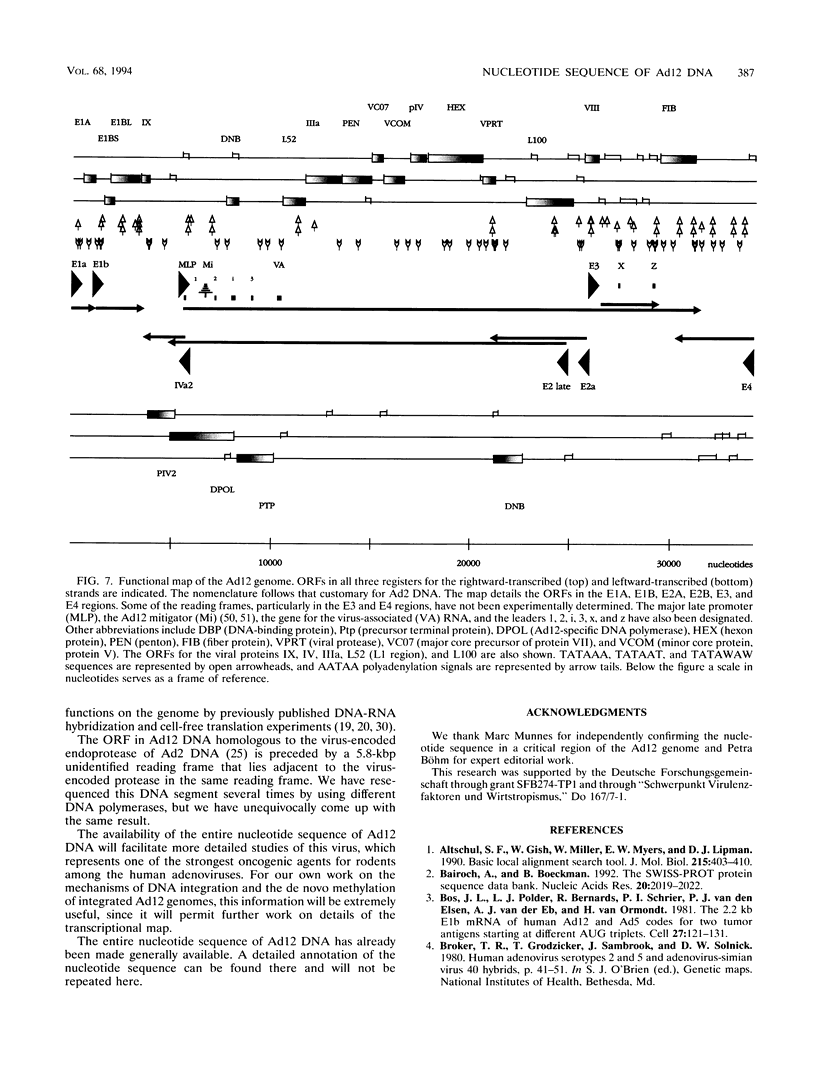
- van Ormondt H., Galibert F. Nucleotide sequences of adenovirus DNAs. Curr Top Microbiol Immunol. 1984;110:73–142. doi: 10.1007/978-3-642-46494-2_4. [DOI] [PubMed] [Google Scholar]


