Abstract
Proton/sulfate cotransporters in the plasma membranes are responsible for uptake of the environmental sulfate used in the sulfate assimilation pathway in plants. Here we report the cloning and characterization of an Arabidopsis thaliana gene, AST68, a new member of the sulfate transporter gene family in higher plants. Sequence analysis of cDNA and genomic clones of AST68 revealed that the AST68 gene is composed of 10 exons encoding a 677-aa polypeptide (74.1 kDa) that is able to functionally complement a Saccharomyces cerevisiae mutant lacking a sulfate transporter gene. Southern hybridization and restriction fragment length polymorphism mapping confirmed that AST68 is a single-copy gene that maps to the top arm of chromosome 5. Northern hybridization analysis of sulfate-starved plants indicated that the steady-state mRNA abundance of AST68 increased specifically in roots up to 9-fold by sulfate starvation. In situ hybridization experiments revealed that AST68 transcripts were accumulated in the central cylinder of sulfate-starved roots, but not in the xylem, endodermis, cortex, and epidermis. Among all the structural genes for sulfate assimilation, sulfate transporter (AST68), APS reductase (APR1), and serine acetyltransferase (SAT1) were inducible by sulfate starvation in A. thaliana. The sulfate transporter (AST68) exhibited the most intensive and specific response in roots, indicating that AST68 plays a central role in the regulation of sulfate assimilation in plants.
In higher plants, sulfur metabolism is initiated by the uptake of sulfate by roots from the environment. Plants assimilate inorganic sulfate into Cys, the first sulfur-containing amino acids, and various sulfur-containing secondary metabolites. Thus, plants serve as nutritional sulfur sources for animals (1). Uptake of sulfate by plants is considered to be the key entry step of the sulfur cycle in the nature. Because the sulfate transporter protein is involved in this initial step, it may play a central role in the regulation of the entire sulfur metabolism pathway by controlling the import of available sulfate. As yet, no detailed molecular biological investigation has been carried out for this important process.
Isolation of cDNA clones encoding sulfate transporters has been recently reported for tropical legume Stylosanthes hamata (2), Arabidopsis thaliana (3), and barley (Hordeum vulgare) (accession no. U52867). From our recent studies (3), there were at least three different sulfate transporter homologues in A. thaliana showing different expression patterns. After entry into root cells, sulfate is delivered to various parts of tissues through the vascular system. The process of “long-distance translocation” (4) of sulfate may require several types of transporters responsible for cell-to-cell movement of sulfate across the plasma membrane. Loading of sulfate into the vascular tissues in roots and unloading of sulfate into the leaf cells are assumed to be the two essential steps in this process. In the present study, we have shown that these two events are possibly controlled by the same sulfate transporter gene, AST68, in A. thaliana.
With respect to the regulation of the entire sulfate assimilation pathway, fluctuation of the extracellular sulfate concentration may act as a signal for the modulation of gene expression. From the earlier physiological experiments on membrane vesicles (5) and cell cultures (6), it is demonstrated that plants can adapt to low sulfate availability by modulating the sulfate transport activity. Because nearly all the genes for the sulfate assimilation enzymes of A. thaliana have been cloned or reported in the expressed sequence tag (EST) database, it is now quite possible to determine which proteins or enzymes in the Cys biosynthesis are regulated. The present study is the first report of a complete analysis of the regulation of the sulfate assimilation pathway in plants. The results show which of the biosynthetic steps are regulated at the level of mRNA expression during adaptation to sulfate deficiency.
MATERIALS AND METHODS
Plant Materials and Yeast and Bacterial Strains.
A. thaliana ecotype Columbia was grown on germination medium (GM) agar medium (7) at 22°C under 16-h/8-h light and dark cycles. Three-week-old plants were subjected to sulfate starvation for 2 days on the sulfate-deficient GM agar medium in which the sulfate salts were replaced with equivalent amounts of chloride salts. As a control experiment, plants were transferred to the fresh GM agar medium, which was subsequently cultured for 2 days. For complementation studies, S. cerevisiae strain YSD1 (Matα, his3-Δ1, leu2, trp1–289, ura3–52, sul1), a disruption mutant of the yeast sulfate transporter gene SUL1 was provided by M. J. Hawkesford (University of Bristol, U.K.) (8). E. coli strains Y1088 (supE, supF, metB, trpR, hsdR−, hsdM+, tonA21, strA, ΔlacU169, mcrA, proC::Tn5/pMC9), K802 (galK2, galT22, hsdR2(rk−, mk+), lacY1, mcrA−, mcrB−, metB1, mrr+, supE44) and XL1-Blue [endA1, gyrA96, hsdR17, lac−, recA1, relA1, supE44, thi-1, (F′ lacqZΔM15, proAB, Tn10)] were used for propagation of λgt11, λEMBL3, and plasmid DNAs, respectively.
Isolation of cDNA and Genomic Clones.
Approximately 5.0 × 105 amplified plaques of the λgt11 cDNA library of A. thaliana ecotype Columbia (3) were screened with the 32P-labeled cDNA insert of an A. thaliana EST, 142F20T7 (accession no. T76088) (9). Hybridization of the membranes (Hybond N+, Amersham) was carried out at 65°C in 5× SSPE (0.9M NaCl/0.05 M sodium phosphate, pH 7.7/5 mM EDTA), 0.5% SDS, 5× Denhardt’s solution, and 20 mg/liter salmon sperm DNA. Final washing of the membranes was conducted at 65°C in 0.1× SSPE and 0.1% SDS (10). For isolation of genomic clones, approximately 1.0 × 105 amplified plaques of the Arabidopsis λEMBL3 genomic library (CLONTECH) were screened with the 32P-labeled cDNA insert of AST68. Hybridization and washing of the membranes (Hybond N+, Amersham) were carried out in the same condition as described for the screening of the λgt11 cDNA library.
DNA Sequencing and Determination of the Transcriptional Start Point.
cDNA and genomic fragments of the isolated clones were cloned in the appropriate cloning sites of pBluescript II SK(−) (Stratagene). All clones were sequenced on both strands using a series of overlapping exonuclease III digested clones created with the Exo/Mung deletion kit (Stratagene). Sequencing was carried out by the dideoxy-chain termination method using Thermo Sequenase (Amersham) and a Shimadzu DNA sequencer model DSQ1000. The primer extention experiment was carried out as described in ref. 11 with some modification. Total RNA (20 μg) from A. thaliana leaves was annealed with the synthetic oligonucleotide (10 pmol, 5′-TGAATATAAAATGTCAGGGA-3′) prepared at the position of 220 nucleotides upstream of the translational initiation codon. Reverse transcription was carried out by 40 units of Moloney murine leukemia virus reverse transcriptase (United States Biochemical) at 37°C in the presence of 0.1 mM of dNTP and 1.85 MBq of [α-32P]dCTP. The extention product was separated in a 5% polyacrylamide sequencing gel, and the signal was detected by the BAS-2000 image analyzer (Fuji). Sequencing products of M13mp18 with the M13-P4 oligonucleotide primer were used as size markers.
Complementation of a Yeast Mutant.
The coding sequence of the Arabidopsis cDNA clone AST68 was amplified by PCR as a BamHI-ended DNA fragment with a set of synthetic oligonucleotide primers (5′-CTTGGATCCATGAAAGAGAGAGATTCAGAG-3′ and 5′-ATTGGATCCTGGTCCTTTGAAAACTGTTTC-3′), using Pfu DNA polymerase (Stratagene). The amplified fragment was inserted in the BamHI site of an yeast expression vector, pYES2 (Invitrogen), under control of the GAL1 promoter. The resultant plasmid, pYAT68, was used for the transformation of YSD1 (8) by the electrotransformation method (12). Transformants were replica plated onto synthetic minimal media (13) containing galactose (20 g/liter) as a carbon source and 0.1 mM MgSO4 as a sulfur source (8) and incubated at 30°C.
Southern Hybridization Analysis.
Genomic DNA was isolated from the leaves of 3-week-old A. thaliana plants (14). Restriction fragment length polymorphism (RFLP) mapping of AST68 was carried out by the 30 recombinant inbred (RI) lines (15). Genomic DNA (5 μg) was digested with restriction enzymes, separated in a 0.7% agarose gel, and transferred to a Hybond N+ membrane (Amersham). DNA blots were probed with the 32P-labeled full-length cDNA fragment of AST68. Hybridization and washing of the membranes were carried out as described for library screening. Hybridization signals were detected by the BAS-2000 image analyzer (Fuji). The map distance was calculated by C. Lister (John Innes Institute, Norwich, U.K.), based on the RFLP patterns in the RI lines generated by DraI digestion of genomic DNAs.
Northern Hybridization Analysis.
Total RNA was isolated from the leaves and roots of 3-week-old A. thaliana plants by a phenol/SDS method and precipitated by LiCl as described in ref. 10. For RNA blot analyses, 20 μg of total RNA was separated under denaturing conditions in a 1.0% agarose gel containing formaldehyde and transferred to Hybond N+ membranes (Amersham). RNA blots were hybridized with 32P-labeled probe DNAs synthesized from cDNA fragments AST68 (this study), AST56 (3), EST-76E7T7 (accession no. T21459) (9), APS1 (16), ASA1 (17) (identical with APS2) (18), APK1 (19) (identical with ATG1/1) (20), APR1 (21) (identical with PRH-19) (22), SAL1 (23), SIR (24), OAS-TL 5–8 (25) (identical with AT-CYS-3A) (26), OAS-TL 7–4 (25), SAT1 (27) (identical with SAT5) (28), SAT-A (29) (identical with Sat-1) (30), and SAT52 (accession no. U30298). The EST clone, 201K3T7 (accession no. H77005) (9), which is identical with SIR (24), was obtained from the Arabidopsis Biological Resource Center of Ohio State University. cDNA fragments of SAL1 (23) and SAT52 were amplified from the cDNA library by PCR with synthetic oligonucleotides prepared according to the reported nucleotide sequence in the database. OAS-TL 5–8 (25), OAS-TL 7–4 (25), and SAT-A (29) were provided by R. Hell (Ruhr-Universität Bochum, Germany). ASA1 (17) was provided by J.-C. Davidian (Ecole Nationale Superieure Agronomique de Montpellier, France). Relative values of mRNA transcripts were calculated based on the hybridization intensities of specific signals on the blot quantified by the BAS-2000 image analyzer (Fuji). To verify equivalent loadings of RNA on blots, membranes were probed with a 32P-labeled rice rDNA (pRR217) (31). Hybridization and washing conditions were the same as described above.
In Situ Hybridization.
In situ hybridization experiments were carried out on A. thaliana and radish. Thirteen-day-old A. thaliana and 5-day-old radish were transferred to sulfate-deficient medium for 2 days. Radish and A. thaliana seedlings grown in sulfate-starved and control conditions were fixed in 4% formaldehyde, 50% ethanol, and 5% acetic acid for 3 hr at room temperature. Fixed tissues were dehydrated and embedded in paraffin according to standard procedures. Ten-micrometer sections were mounted onto slides coated with 3-aminopropyltriethoxysilane and pretreated for hybridization according to Angerer and Angerer (32). 35S-UTP-labeled sense and antisense mRNA were generated by run-off transcription with T7 and T3 RNA polymerase (Promega). Labeled mRNA probes were hydrolyzed to an average length of 300 nt (32). The hybridization mix contained 35S-labeled mRNA (5 × 106 cpm per slides), 10 mM Tris⋅HCl (pH8.5), 50% formamide, 0.3M NaCl, 1 mM EDTA, 150 μg ml−1 yeast tRNA, 1× Denhardt’s, 10% dextran sulfate, and 70 mM DTT. RNase treatment, washing steps, coating with Kodak NBT2 emulsion, and the development of slides were performed as described in ref. 32. Sections were stained with toluidine blue, dehydrated, and mounted in mounting medium (Depex). Photographs were taken in a Diaplan microscope (Leica) using dark-field optics (Fuji ASA 100 film).
RESULTS
Cloning and Functional Identification of a Sulfate Transporter Gene, AST68.
An Arabidopsis EST clone, 142F20T7 (accession no. T76088) (9), exhibiting high sequence similarity with the sulfate transporters of the tropical legume S. hamata (2), was used as a probe for isolation of a full-length clone from a λgt11 cDNA library. The isolated cDNA clone, AST68, contained a 2.4-kb-length insert that revealed to have an ORF encoding a polypeptide of 677 aa (Fig. 1B). The presence of an in-frame TAG termination codon 39 nucleotides upstream of the translational initiation codon ensured that this ORF encodes the full coding region of the AST68 polypeptide.
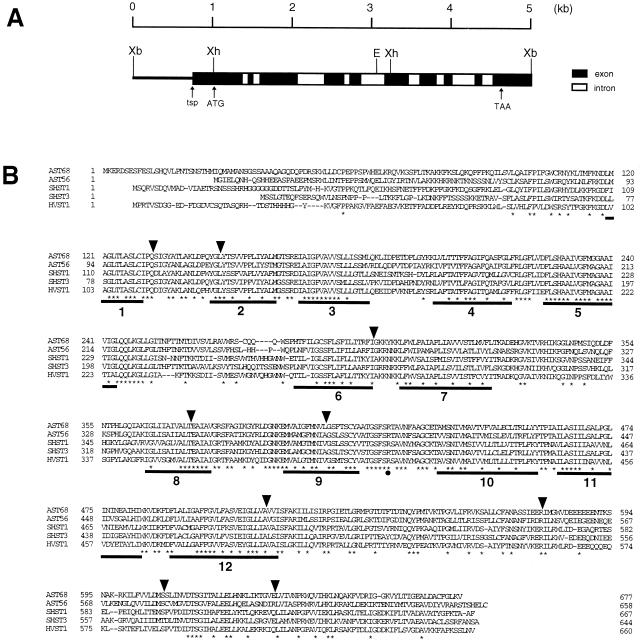
Figure 1.
Structure of the AST68 gene. (A) The partial restriction map of the AST68 genomic region. Solid bars and open bars indicate exons and introns, respectively. The transcriptional starting point (tsp) and the translational initiation (ATG) and termination (TAA) codons are indicated by arrows. Abbreviations of restriction enzymes are as follows: E, EcoRI; Xb, XbaI; Xh, XhoI. (B) Alignment of the deduced amino acid sequences of the plant sulfate transporters: Arabidopsis AST68, Arabidopsis AST56 (3), Stylosanthes hamata SHST1, SHST3 (2), Holdeum vulgare HVST1 (accession no. U52867). Consensus amino acids are indicated by asterisks. Gaps in the sequence inserted to obtain the best alignment are indicated by dashes. Insertion sites of the introns of the AST68 gene are indicated by arrowheads. Solid bars indicate 12 putative MSDs. A conserved basic amino acid residue (Arg-407) between MSDs 9 and 10 is indicated by a solid circle.
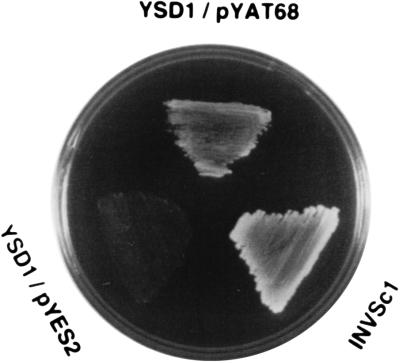
The sulfate uptake function of the AST68 polypeptide was tested in the yeast sulfate transporter mutant strain YSD1 (8). Expression of AST68 allowed YSD1 to grow on the minimal medium containing 0.1 mM of sulfate as a sole sulfur source only in the presence of galactose (Fig. 2). The strain was unable to grow with glucose (not shown). These data confirmed that AST68 encodes a functional homologue of a yeast sulfate transporter (8).
Figure 2.
Complementation analysis of AST68. A wild-type strain, INVSc1, and the sulfate transporter-deficient mutant, YSD1, transformed with vectors pYES2 (control) and pYAT68 (harboring the AST68 coding region), were grown at 30°C on the synthetic minimal media containing galactose (20 g/liter) as a carbon source and 0.1 mM MgSO4 as a sulfur source as described in ref. 9.
A genomic clone corresponding with the cDNA clone, AST68, was isolated from a λEMBL3 library. Several clones were isolated that hybridized with AST68. Sequence analysis of a 5-kb XbaI fragment (Fig. 1A) from one of these clones revealed that the AST68 gene consists of 10 exons and 9 introns. All of the exon/intron junctions had the consensus GT/AG splice donor and acceptor sites. Primer extension experiments were carried out with a synthetic oligonucleotide primer prepared according to the sequence of the AST68 genomic clone. The transcriptional start point (tsp) was located 285 nucleotides upstream of the translational initiation codon (Fig. 1A). The 5′ untranslated region between the tsp and the 5′ end of the cDNA clone was amplified by reverse transcriptase–PCR (RT-PCR) for the determination of the nucleotide sequence.
Predicted Structure of the AST68 Polypeptide.
The hydropathy profile of AST68 predicted by the toppredii program (33) indicated the presence of 12 hydrophobic membrane spanning domains (MSD). This structural feature was well conserved in eukaryotic membrane-bound transporter proteins. According to this model, several basic amino acid residues (Lys, Arg) were located on both sides of the membrane. Among them, Arg-407, which is located between MSDs 9 and 10, was the only basic residue identical to the other known eukaryotic sulfate transporters (Fig. 1B). As previously indicated by Smith et al. (2), this residue may have some functional significance for binding of the sulfate anion on the membrane surface. The calculated model also predicted the putative N-glycosylation site at Asn-255 between MSDs 5 and 6 to be located on the extracellular side.
Phylogenic Relationship.
Similarities between the amino acid sequence and other eukaryotic sulfate transporters were calculated by the genetyx program (Software Development, Tokyo) as follows: S. cerevisiae Sul1, 28% (accession no. X82013) (8); N. crassa Cys14, 21% (accession no. M59167) (34); S. hamata Shst1, 49% (accession no. X82255) (2); S. hamata Shst2, 49% (accession no. X82256) (2); S. hamata Shst3, 64% (accession no. X82254) (2); A. thaliana AST56, 63% (accession no. D85416) (3); soybean nodule GmN#70, 54% (accession no. D13505) (35); H. vulgare Hvst1, 51% (accession no. U52867); mouse, 28% (accession no. D42049); human DTD, 28% (accession no. U14528) (36); rat Sat-1, 27% (accession no. L23413) (37); and human DRA, 25% (accession no. L02785) (38). The phylogenic relationship of these amino acid sequences indicated that the two sulfate transporters of A. thaliana, AST56 (3) and AST68 (this study), fall into a group that includes the plant low-affinity transporters. In spite of the structural and functional similarity, AST68 is highly divergent from the yeast sulfate transporter, SUL1 (8).
Chromosomal Location of the AST68 Gene.
Southern blot analysis of the AST68 gene was carried out for the A. thaliana genomic DNA digested with several restriction enzymes. Restriction with BamHI, EcoRV, SacI, and XbaI gave a single hybridization signal under high stringency washing conditions. These data indicated that AST68 was a single-copy gene in the A. thaliana genome. Chromosomal location of the AST68 gene was calculated from the polymorphism in DraI digestion of 30 recombinant inbred lines as described in ref. 15. The AST68 gene was mapped within the top of chromosome 5 between the two markers, g3837 and g4560.
In Situ Localization of the AST68 Transcripts.
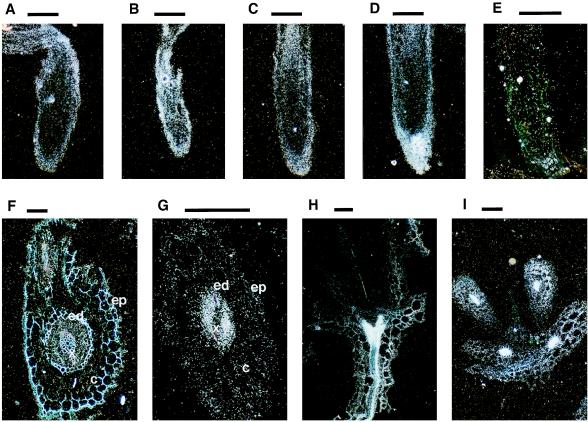
In situ hybridization on longitudinal sections of A. thaliana roots showed that AST68 transcripts are accumulated in the root cap (Fig. 3A) and are strongly induced by sulfate starvation (Fig. 3B). Heterologous hybridization on longitudinal sections of radish roots showed the same distribution pattern as in the case of A. thaliana (Fig. 3 C and D). No signals were detected by hybridization with the sense probe (Fig. 3E). To identify the cell-specific accumulation of the AST68 transcripts, hybridization was carried out on cross-sections of radish roots. Positive signals could be detected particularly inside the central cylinder. However, no apparent signals were observed in the xylem, endodermis, cortex, and epidermis. The cross-section in Fig. 3F includes the lateral and primary roots. Similar distribution patterns of the AST68 transcripts were observed in these two different stages of root elongation. Positive signals localized in the central cylinder were strongly induced by sulfate starvation (Fig. 3G). Hybridization on longitudinal and cross-sections of A. thaliana shoots indicated that AST68 transcripts are also localized in the vascular tissues of aerial parts (Fig. 3 H and I). However, these signals in shoots did not change by sulfate starvation (data not shown). These data suggested that a single sulfate transporter, AST68, may participate in symplastic translocation of sulfate in the central cylinder both in roots and leaves.
Figure 3.
In situ hybridization analysis of the AST68 gene. Longitudinal sections of A. thaliana roots in the normal condition (A) and in the sulfate-starved condition (B). Longitudinal sections of radish roots in the normal condition (C) and in the sulfate-starved condition (D). A longitudinal section of radish roots in the sulfate-starved condition hybridized with the sense probe (E). Cross-sections of radish roots in the normal condition (F) and in the sulfate-starved condition (G). A longitudinal section of an A. thaliana shoot in the normal condition (H). A cross-section of an A. thaliana shoot in the normal condition (I). Bars = 100 μm. c, cortex; ed, endodermis; ep, epidermis; x, xylem.
Regulation of Genes Involved in Sulfate Assimilation.
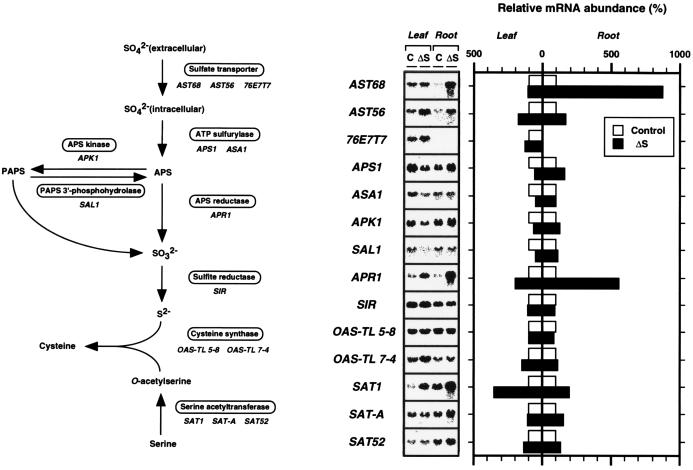
Expression of genes for sulfate assimilation enzymes was analyzed in A. thaliana plants grown with sulfate and without sulfate for 2 days (Fig. 4). Three genes, AST68, APR1, and SAT1, were inducible by the short-term sulfate starvation among the 14 genes examined in our experiments. The mRNA abundance of AST68 increased up to ca. 9-fold in roots by sulfate starvation. The expression of the minor isoform, AST56 (3), was also enhanced in roots to some extent (1.5- to 2-fold) by the same treatment. However, the transcript levels of AST68 and the leaf specific homologue 76E7T7 (accession no. T21459) were not affected by sulfate deprivation in leaves. APR1 (21, 22), recently identified to encode an APS reductase isoform responsible for the reduction of adenosine 5′-phosphosulfate (APS) to sulfite, increased ca. 5.5-fold in roots and 2-fold in leaves. SAT1 (27), which encodes a serine acetyltransferase (SAT) isoform and catalyzes the formation of O-acetylserine from Ser, increased ca. 3.5-fold in leaves and 2-fold in roots. It is notable that the expression of SAT-A (26) and SAT52 (accession no. U30298), which encode the other two SAT isoforms, exhibited no changes in response to sulfate deprivation. This is also true of the serine acetyl transferase SAT2 gene from watermelon (39).
Figure 4.
Northern blot analysis of total RNA of A. thaliana. The name of the enzyme involved in each biosynthetic step and the corresponding A. thaliana cDNA clones used in this experiment are noted. Total RNA was extracted from leaves and roots of the plants grown under a control condition (C) and a sulfate-starved condition (ΔS). Three-week-old A. thaliana plants grown on GM-agar medium (MS salts, 1% sucrose; ref. 8) were transferred to sulfate-deficient GM-agar medium (MS salts as chloride instead of sulfate, 1% sucrose), which was subsequently grown for 2 days. Total RNA (20 μg) was separated in a 1.0% agarose gel containing formaldehyde, transferred to nylon membranes (Hybond N+, Amersham), and hybridized with the 32P-labeled cDNA inserts. Washing of the membranes was carried out under the same conditions as described in Fig. 4. Relative mRNA abundance was quantified by the hybridization intensities of the blots using the BAS-2000 image analyzer (Fuji). Transcript levels changed by sulfate starvation are shown in comparison with those in the control condition (100%) of leaves and roots, respectively.
With respect to the other genes responsible for Cys biosynthesis, we could find no significant up-regulation by sulfate starvation, as previous studies have partially suggested (40). The mRNA expression of ATP sulfurylase and APS kinase was down-regulated by sulfate starvation in leaves. Expression of the halotorerant gene SAL1, which was characterized to be involved in dephosphorylation of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to APS (22), was also depressed in sulfate-starved leaves. From these results, it was indicated that the expression of the genes involved in sulfate assimilation is not coordinately regulated by sulfate starvation in plants. We could also indicate that sulfate transporter AST68 was the most prominently responding gene in the Cys biosynthetic pathway under sulfate-deprived conditions.
DISCUSSION
Through combined analyses of the EST database and RNA blot hybridization, it is likely that there are at least three different types of sulfate transporters in A. thaliana. Among them, the mRNA expression of AST68 is strongly induced in roots, but not in leaves, by sulfate starvation (Fig. 4). In situ hybridization analyses revealed that AST68 is expressed in the root cap and in the central cylinder of roots and leaves. We have also shown that AST68 is induced by sulfate starvation specifically in roots. Expression and induction of the AST68 transcripts in the central cylinder of roots were limited in the cell layers between the endodermal cell and the xylem vessel (Fig. 3). Endodermal cells form an impermeable cell wall, collectively termed the Casparian strips, and serve to limit the apoplastic diffusion of solutes into the root vascular system. Although the mRNA levels may not necessarily correspond to the protein levels, up-regulation and accumulation of the AST68 transcripts found in the central cylinder may enhance the efficiency of symplastic sulfate translocation from the endodermal cell toward the xylem vessel responding to sulfate starvation. Alternatively, induction of AST68 was caused by the requirement of sulfate to synthesize sulfur-containing metabolites in these cells.
The next topic of the present study focused on the mRNA expression of APS reductase by nutritional sulfur stress. Recently, Setya et al. (21) and Gutierrez-Marcos et al. (22) demonstrated the existence of APS reductase in the higher plant A. thaliana. The existence of a PAPS-dependent sulfate-reduction pathway (41) remains unresolved. Based on the current information, it appears that APS kinase, the enzyme catalyzing the phosphorylation of APS to PAPS, is probably not directly involved in the reductive sulfate assimilation pathway. Rather, it serves the branch pathway leading to sulfation of various plant metabolites such as sulfated flavonols (42), sulfolipids (43), and glucosinolates (44). Thus, APS serves as a central branch point intermediate both for the sulfate reduction and sulfation pathways. APS reductase and APS kinase must therefore compete for their common substrate, and this may serve in regulation. At the level of mRNA abundance, the expression of APR1 was enhanced by sulfate starvation, whereas the expression of APK1 was down-regulated in leaves. These results suggested that the overaccumulation of the APR1 transcripts is necessary to increase the efficiency of Cys biosynthesis from limited amounts of sulfate. It is reported that APS sulfotransferase activity is increased by sulfate starvation in water lentil (Lemna minor) (45). Our findings on regulation of APR1 may describe that the enhancement of APS sulfotransferase activity is closely related to the accumulation of mRNA. Determination of the enzyme activity in A. thaliana is required to explain the functional participation of the APR1 gene product in enhancement of sulfate reduction and Cys biosynthesis. In our experiments, mRNA expression of PAPS 3′-phosphohydrolase (SAL1), which may participate in a “futile cycle” of APS and PAPS, was down-regulated as in the case of APK1. It apparently shows the coordinated regulation of PAPS 3′-phosphohydrolase with APS reductase and APS kinase at the mRNA levels. However, the SAL1 enzyme is putatively localized in the cytoplasm. Thus, it may have no correlation with the other two chloroplast-localizing enzymes.
With respect to the mRNA expression of three serine acetyltransferase isoforms, SAT1 was the only one to be induced by sulfate starvation. The two sequential steps to synthesize Cys from Ser is catalyzed by the multi-enzyme complex of serine acetyltransferase (SAT) and cysteine synthase (CS). This may prevent the diffusion of the intermediate substrate, O-acetylserine (OAS) (46). It can be assumed that SAT1 was induced to couple with the excess amounts of the counterpart CS isoform to obtain the full enzyme activity to synthesize Cys from reduced amounts of sulfide under sulfate-starved conditions. Our previous studies on transgenic tobaccos overexpressing a spinach CS isoform indicated that addition of OAS and sulfite or sulfide enhanced the formation of Cys in isolated chloroplasts (47). These data support an idea that induction of SAT1 together with APR1 may achieve the overaccumulation of OAS and sulfite required for Cys biosynthesis. The alternative explanation is that overproduction of OAS by SAT1 is required for the regulation of sulfate assimilation as has been reported for the induction of the APS sulfotransferase activity by the addition of OAS in water lentil (48). Determination of the SAT1 protein level under sulfate-starved conditions and identification of the counterpart CS isoform should be further investigated to explain the involvement of SAT1 gene expression in the enhancement of Cys biosynthesis.
Our present study demonstrated that the regulation system of sulfate assimilation in plants is definitely different from those of yeast (49) and bacteria (50); in plants, only restricted numbers of genes were induced in response to the sulfate-deficient stress. The difference in gene expression may explain the evolution process that the plants have experienced, acquiring suitable systems to survive in various environmental conditions and to contribute in the global cycle of sulfur in nature.
Acknowledgments
We thank the Arabidopsis Biological Resource Center of Ohio State University for EST clones 76E7T7, 142F20T7, and 201K3T7; Dr. R. Hell (Ruhr-Universität Bochum, Germany) for cDNA clones OAS-TL 5–8, OAS-TL 7–4, and SAT-A; Dr. J.-C. Davidian (Ecole Nationale Superieure Agronomique de Montpellier, France) for cDNA clone ASA1; Dr. M. J. Hawkesford (University of Bristol, U.K.) for S. cerevisiae strain YSD1; and Dr. C. Lister (John Innes Institute, Norwich, U.K.) for the mapping of the AST68 gene. This research was supported in part by grants-in-aid for scientific research from Ministry of Education, Science and Culture, Japan, and by Research for the Future Program (96I00302) from the Japan Society for the Promotion of Science. T. L. was supported by the National Science Foundation.
ABBREVIATIONS
- APS
adenosine 5′-phosphosulfate
- CS
cysteine synthase
- EST
expressed sequence tag
- MSD
membrane spanning domain
- OAS
O-acetylserine
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
- SAT
serine acetyltransferase
- GM
germination medium
Footnotes
References
- 1.Giovanelli J, Mudd S H, Datko A N. In: The Biochemistry of Plants: A Comprehensive Treatise, Vol. 5: Amino Acids and Derivatives. Stumpf P K, Conn E E, editors. New York: Academic; 1980. pp. 453–505. [Google Scholar]
- 2.Smith F W, Ealing P M, Hawkesford M J, Clarkson D T. Proc Natl Acad Sci USA. 1995;92:9373–9377. doi: 10.1073/pnas.92.20.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi H, Sasakura N, Noji M, Saito K. FEBS Lett. 1996;392:95–99. doi: 10.1016/0014-5793(96)00787-9. [DOI] [PubMed] [Google Scholar]
- 4.Clarkson D T, Hawkesford M J, Davidian J-C. In: Sulfur Nutrition and Assimilation in Higher Plants. De Kok L J, Stulen I, Rennenberg H, Brunold C, Rauser W E, editors. The Hague: SPB Academic Publishing; 1993. pp. 3–19. [Google Scholar]
- 5.Hawkesford M J, Davidian J-C, Grignon C. Planta. 1993;190:297–304. [Google Scholar]
- 6.Smith I K. Plant Physiol. 1975;55:303–307. doi: 10.1104/pp.55.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valvekens D, Van Montagu M, Van Lijsebettens M. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith F W, Hawkesford M J, Prosser I M, Clarkson D T. Mol Gen Genet. 1995;247:709–715. doi: 10.1007/BF00290402. [DOI] [PubMed] [Google Scholar]
- 9.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, Retzel E, Somerville C. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 11.Mcknight S L, Kingsbury R. Science. 1982;217:316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- 12.Becker D M, Guarente L. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 13.Sherman F. Methods Enzymol. 1991;194:4–21. doi: 10.1016/0076-6879(91)94005-w. [DOI] [PubMed] [Google Scholar]
- 14.Dellaporta S L, Wood J, Hicks J B. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- 15.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 16.Leustek T, Murillo M, Cervantes M. Plant Physiol. 1994;105:897–902. doi: 10.1104/pp.105.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan H M, Cathala N, Grignon C, Davidian J-C. J Biol Chem. 1996;271:12227–12233. doi: 10.1074/jbc.271.21.12227. [DOI] [PubMed] [Google Scholar]
- 18.Murillo M, Leustek T. Arch Biochem Biophys. 1995;323:195–204. doi: 10.1006/abbi.1995.0026. [DOI] [PubMed] [Google Scholar]
- 19.Jain A, Leustek T. Plant Physiol. 1994;105:771–772. doi: 10.1104/pp.105.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arz H E, Gisselmann G, Schiffmann S, Schwenn J D. Biochim Biophys Acta. 1994;1218:447–452. doi: 10.1016/0167-4781(94)90203-8. [DOI] [PubMed] [Google Scholar]
- 21.Setya A, Murillo M, Leustek T. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez-Marcos J F, Roberts M A, Campbell E I, Wray J L. Proc Natl Acad Sci USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintero F J, Garciadeblás B, Rodríguez-Navarro A. Plant Cell. 1996;8:529–538. doi: 10.1105/tpc.8.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brühl A, Haverkamp T, Gisselmann G, Schwenn J D. Biochim Biophys Acta. 1996;1295:119–124. doi: 10.1016/0167-4838(96)00066-0. [DOI] [PubMed] [Google Scholar]
- 25.Hell R, Bork C, Bogdanova N, Frolov I, Hauschild R. FEBS Lett. 1994;351:257–262. doi: 10.1016/0014-5793(94)00872-8. [DOI] [PubMed] [Google Scholar]
- 26.Barroso C, Vega J M, Gotor C. FEBS Lett. 1995;363:1–5. doi: 10.1016/0014-5793(95)00255-8. [DOI] [PubMed] [Google Scholar]
- 27.Murillo M, Foglia R, Diller A, Lee S, Leustek T. Cell Mol Biol Res. 1995;41:425–433. [PubMed] [Google Scholar]
- 28.Ruffet M-L, Lebrun M, Droux M, Douce R. Eur J Biochem. 1995;227:500–509. doi: 10.1111/j.1432-1033.1995.tb20416.x. [DOI] [PubMed] [Google Scholar]
- 29.Bogdanova N, Bork C, Hell R. FEBS Lett. 1995;358:43–47. doi: 10.1016/0014-5793(94)01392-e. [DOI] [PubMed] [Google Scholar]
- 30.Roberts M A, Wray J L. Plant Mol Biol. 1996;30:1041–1049. doi: 10.1007/BF00020814. [DOI] [PubMed] [Google Scholar]
- 31.Takaiwa F, Oono K, Sugiura M. Plant Mol Biol. 1985;4:355–364. doi: 10.1007/BF02418257. [DOI] [PubMed] [Google Scholar]
- 32.Angerer L M, Angerer R C. In: In Situ Hybridization: A Practical Approach, The Practical Approach Series. Wilkinson D G, editor. Oxford: IRL Press; 1992. pp. 15–32. [Google Scholar]
- 33.Claros M G, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 34.Ketter J S, Jarai G, Fu Y-H, Marzluf G A. Biochemistry. 1991;30:1780–1787. doi: 10.1021/bi00221a008. [DOI] [PubMed] [Google Scholar]
- 35.Kouchi H, Hata S. Mol Gen Genet. 1993;238:106–109. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 36.Hätbacka J, de la Chapelle A, Mahtani M M, Clines G, Reeve-Daly M P, Daly M, Hamilton B A, Kusumi K, Trivedi B, Weaver A, Coloma A, Lovett M, Buckler A, Kaitila I, Lander E S. Cell. 1994;78:1073–1087. doi: 10.1016/0092-8674(94)90281-x. [DOI] [PubMed] [Google Scholar]
- 37.Bissig M, Hagenbuch B, Stieger B, Koller T, Meier P J. J Biol Chem. 1994;269:3017–3021. [PubMed] [Google Scholar]
- 38.Schweinfest C W, Henderson K W, Suster S, Kondoh N, Papas T S. Proc Natl Acad Sci USA. 1993;90:4166–4170. doi: 10.1073/pnas.90.9.4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saito K, Inoue K, Fukushima R, Noji M. Gene. 1996;189:57–63. doi: 10.1016/s0378-1119(96)00833-5. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi H, Saito K. Plant Physiol. 1996;112:273–280. doi: 10.1104/pp.112.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hell R. Planta. 1997;202:138–148. doi: 10.1007/s004250050112. [DOI] [PubMed] [Google Scholar]
- 42.Lacomme C, Roby D. Plant Mol Biol. 1996;30:995–1008. doi: 10.1007/BF00020810. [DOI] [PubMed] [Google Scholar]
- 43.Kleppinger-Sparace K F, Mudd J B. Plant Physiol. 1990;93:256–263. doi: 10.1104/pp.93.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schnug E. In: Sulfur Nutrition and Assimilation in Higher Plants. De Kok L J, Stulen I, Rennenberg H, Brunold C, Rauser W E, editors. The Hague: SPB Academic Publishing; 1993. pp. 179–190. [Google Scholar]
- 45.Brunold C, Suter M, Lavancy P. Physiol Plant. 1987;70:168–174. [Google Scholar]
- 46.Saito K, Yokoyama H, Noji M, Murakoshi I. J Biol Chem. 1995;270:16321–16326. doi: 10.1074/jbc.270.27.16321. [DOI] [PubMed] [Google Scholar]
- 47.Saito K, Kurosawa M, Tatsuguchi K, Takagi Y, Murakoshi I. Plant Physiol. 1994;106:887–895. doi: 10.1104/pp.106.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuenschwander U, Suter M, Brunold C. Plant Physiol. 1991;97:253–258. doi: 10.1104/pp.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuras L, Thomas D. FEBS Lett. 1995;367:15–18. doi: 10.1016/0014-5793(95)00528-h. [DOI] [PubMed] [Google Scholar]
- 50.Kredich N M. In: Sulfur Nutrition and Assimilation in Higher Plants. De Kok L J, Stulen I, Rennenberg H, Brunold C, Rauser W E, editors. The Hague: SPB Academic Publishing; 1993. pp. 37–47. [Google Scholar]