Abstract
Two RNases H of mammalian tissues have been described: RNase HI, the activity of which was found to rise during DNA replication, and RNase HII, which may be involved in transcription. RNase HI is the major mammalian enzyme representing around 85% of the total RNase H activity in the cell. By using highly purified calf thymus RNase HI we identified the sequences of several tryptic peptides. This information enabled us to determine the sequence of the cDNA coding for the large subunit of human RNase HI. The corresponding ORF of 897 nt defines a polypeptide of relative molecular mass of 33,367, which is in agreement with the molecular mass obtained earlier by SDS/PAGE. Expression of the cloned ORF in Escherichia coli leads to a polypeptide, which is specifically recognized by an antiserum raised against calf thymus RNase HI. Interestingly, the deduced amino acid sequence of this subunit of human RNase HI displays significant homology to RNase HII from E. coli, an enzyme of unknown function and previously judged as a minor activity. This finding suggests an evolutionary link between the mammalian RNases HI and the prokaryotic RNases HII. The idea of a mammalian RNase HI large subunit being a strongly conserved protein is substantiated by the existence of homologous ORFs in the genomes of other eukaryotes and of all eubacteria and archaebacteria that have been completely sequenced.
RNases H are enzymes that specifically degrade the RNA moiety of RNA–DNA hybrids. An enzyme with the specificity of RNase H was first recognized and characterized in extracts of calf thymus tissue (1). Shortly thereafter it was shown that RNase H is not restricted to calf thymus, but that it also exists in prokaryotes (2, 3), other eukaryotes (4–6), and retroviruses (7). These early studies not only pointed to the ubiquitous occurrence of RNase H and therefore its possible important role in nucleic acid metabolism, but they also indicated the existence of multiple forms of RNases H. Two mammalian RNases H were described that can be differentiated by physical, biochemical, and serological parameters. For bovine RNases H, the larger RNase HI could be correlated with DNA replication and the smaller RNase HII with transcription (8). More recently, RNase HI was discussed in the context of a possible DNA repair function (9) and was shown to participate together with flap endonuclease 1 (FEN1) in the removal of the Okazaki primers during lagging strand DNA synthesis (10). Mammalian RNases H were purified and characterized from a variety of sources, including calf thymus (11), mouse cells (12), Hela cells (13), human placenta (14), and human erythroleukemia cells (9). Recently, the interest in RNases H was roused again, particularly in connection with possible therapeutic implications. An endogenous RNase H activity may play an essential role in biological effects mediated by antisense oligonucleotides (see e.g., ref. 15), molecules considered as potential agents against infectious diseases and pathologies resulting from dysfunctional genes. Extensive knowledge of human RNases H, their structures, catalytic sites, and physiological roles should help with designing selective drugs. Despite the above-indicated importance of elucidating the properties of mammalian RNases H, the best characterized and functionally understood RNases H are the RNase H domains of retroviral reverse transcriptases and the evolutionarily related RNase HI of the eubacterium, Escherichia coli, for both of which crystal structures are available (16, 17). Although the necessity of RNase H activity in connection with reverse transcription of retroviral RNA genomes into double-stranded DNA is intuitively comprehensible, the function of the cellular RNases H is less obvious. If, as suggested earlier (18), “modern” DNA replication evolved from reverse transcription of originally existing RNA genomes into DNA and RNA primers of Okazaki fragments were remnants of this process, the most obvious role for cellular RNases H would be the removal of Okazaki fragments during lagging strand DNA synthesis. In E. coli, however, this task is carried out mainly by DNA polymerase I (19), and the extensively studied RNase HI of this organism was shown to fulfil regulatory functions during the process of genome duplication (20). Much less is known about the second prokaryotic enzyme, RNase HII, which was cloned from E. coli and then was designated a minor activity of unknown function (21). Comparative renaturation gel assay data of E. coli cell extracts (22) and the recently published characterization of the related Streptococcus pneumoniae RNase HII (23) indicate that the importance of these enzymes has been underestimated. This idea recently was corroborated by identification of the homologous gene from the yeast Saccharomyces cerevisiae (24), which in this organism codes for the main RNase H. Here we will present the experimental data for the determination of the complete amino acid sequence of the large subunit of human RNase HI, which will unveil the prokaryotic RNase HII as the evolutionary counterpart of the major mammalian RNase H. We will further show that related ORFs are present in genomes of organisms from all three kingdomes (archaebacteria, eubacteria, and eukaryotes).
MATERIALS AND METHODS
RNase H Purification, Enzyme Activity Assays, and Standard Techniques for Protein Analysis.
Calf thymus RNase HI was purified according to Büsen (25). The RNase H preparation was separated on a preparative 12% SDS/PAGE and stained with Coomassie brilliant blue R-250 (Serva), and the band corresponding to the 32-kDa protein was cut out and concentrated on a specially designed concentration gel according to Vandekerckhove et al. (26). The renaturation gel assay for the in situ detection of RNase H activity was performed according to Frank et al. (22) by using Mn2+ as a divalent cation. Protein concentration was determined by the method of Bradford (27). Discontinuous SDS/PAGE was performed according to Laemmli (28); protein bands were visualized with Coomassie brilliant blue G-250 or by silver staining (29). Western blotting was performed as described (30).
Peptide Sequencing.
Approximately 5 μg of the purified and concentrated bovine 32-kDA polypeptide were blotted from the Vandekerckhove gel onto a poly(vinylidene difluoride) membrane (Millipore) by using a standard blotting tank system. The transfer buffer contained 13 mM sodium carbonate, pH 9.9 and 20% methanol. The protein spot was detected by staining with Coomassie brilliant blue R-250 and afterward excised from the membrane. The blotted protein sample was digested with sequencing grade trypsin (Promega) as described by Fernandez et al. (31), except that the detergent Triton X-100 was replaced by octyl-β-d-glucopyranoside (Boehringer Mannheim). Peptide mapping was carried out on a 1 × 250 mm Vydac C18 column at 40°C with a flow rate of 50 ml/min by using the HP 1100 HPLC system (Hewlett–Packard). Selected peptide fractions were analyzed by automated Edman degradation using the HP G1005A protein-sequencing system (Hewlett–Packard). With the knowledge of the peptide sequences, corresponding human expressed sequence tags (ESTs) were identified in the EST database at the National Center for Biotechnology Information by using blast. This computer analysis enabled us to assemble an ORF coding for around 300 aa and to design primers for the cloning and sequencing of the corresponding cDNA sequence.
PCR Amplification and Cloning Procedures.
To amplify the RNase HI coding sequence, PCRs were performed with Marathon cDNA (CLONTECH) from a human erythroleukemia cell line as a template. The following primers were used for PCR with the Expand PCR system (Boehringer Mannheim): cth5f/EcoRI, derived from an EST sequence (GenBank accession no. H43540), which covers the 5′ end of the ORF, including the ATG start codon (underlined) and introduces an EcoRI site (bold)—5′-GATGTCGGAATTCATGGATCTCAGCGAGCTGGAGAGAGACAATACAGGC-3′, and cth3r/SalI, derived from an EST sequence (EMBL accession no. AA262477), which covers the 3′ end of the ORF, including the TAG stop codon (underlined, complementary strand) and introduces a SalI site (bold)—5′-GATGTCGTCGACTGCTAGAGGCTGGTTGCTGACTCCAGGCCG-3′. A PCR product of about 900 bp in size was obtained, digested with EcoRI and SalI, gel purified, and ligated into an EcoRI/SalI-digested pXa1 expression vector (Boehringer Mannheim). This vector system allows the expression of β-galactosidase fusion proteins after induction with isopropyl-1-thio-β-d-galactopyranoside (IPTG). The ligation sample was transformed into competent XL1 Blue E. coli cells. Single colonies of bacterial cultures were grown in the presence of IPTG and harvested by centrifugation. Bacterial pellets were processed for SDS/PAGE and analyzed on 8.5% gels for the presence of a fusion protein of the correct size (156 kDa).
DNA Sequence Determination.
Nonradioactive DNA sequencing using the alf system (Pharmacia) was performed with eight independently cloned PCR products as templates. Those PCR products were synthesized with the primers cth5f/EcoRI and cth3r/SalI (described above). The following primers were used for the sequencing reactions: forward primer 1, 5′-ATGGATCTCAGCGAGCTGGAG-3′; and reverse primer 1, 5′-CTAGAGGCTGGTTGCTGACTC-3′. To unravel the DNA sequence of the ORF in the region of the start and stop codon, two additional PCR products were synthesized and sequenced. To do that, we again used Marathon cDNA (CLONTECH) from a human erythroleukemia cell as a template and the following primer sets: set 1, forward primer AP1 (suitable for 5′and 3′ rapid amplification of cDNA ends (RACE) with Marathon cDNA (CLONTECH), 5′-GCCCTATAGTGAGTCGTATTAGGATGG-3′, reverse primer cth6/rev, 5′-TTTGCGAAAATGGAGGACACGGACTTTGTC-3′; set 2, forward primer cth2, 5′-CACGTGGAGCCTGTGTTCGGCTTCCCCCAGTTTGTC-3′, reverse primer AP1 [suitable for 5′ and 3′ RACE with Marathon cDNA (CLONTECH)]. With primer set 1 we obtained a PCR product of about 350 bp, and with primer set 2 we got about 480 bp. The PCR product of primer set 1 was sequenced by using the reverse primer cth6/rev, and the product of set 2 was analyzed by using the forward primer cth2. The DNA sequence described in this report was deposited in the GenBank nucleotide sequence database with accession no. Z97029.
Expression, Purification, and Factor Xa Digestion of the β-Galactosidase Fusion Protein.
Recombinant fusion protein from the pXa1 expression plasmid was purified as follows. An overnight culture of 100 ml was grown in Luria broth in the presence of 1 mM isopropyl-1-thio-β-d-galactopyranoside and 25 μg/ml of ampicillin. The cells were harvested by centrifugation and opened by ultrasonic treatment in ice-cold buffer A (PBS + 5 mM EDTA). Then the extract was treated with benzonase (Merck) to destroy RNA and DNA and 8 M urea to solubilize inclusion bodies. The preparation was loaded onto a Bio-Rad 691 prepcell (preparative gel electrophoresis) and separated on a cylindrical 4% gel. Elution fractions were analyzed for the presence of purified β-galactosidase fusion protein by analytical 8.5% SDS/PAGE. The fractions containing the fusion protein were pooled and dialyzed three times against buffer A. Then they were concentrated in the dialysis bag by external treatment with aquacide II (Calbiochem). The concentrated fraction was digested with factor Xa protease at room temperature for different time periods. The efficiency of the digestion was analyzed by 12% SDS/PAGE and Western blotting.
Northern Blot Analysis.
Total RNA from HEK-293 cells (human embryonal kidney cells) was purified with the Trizol kit (Life Technologies, Grand Island, NY) according to the instructions of the manufacturer. This preparation was used for Northern blot analysis. Twenty micrograms of total RNA were electrophoresed in a 1% agarose/formaldehyde gel along with RNA Mr markers and processed for blotting according to standard methods. For the preparation of the probe, a PCR product of 230 bp, located inside the ORF of the human RNase HI large subunit, was randomly labeled by using the Digoxigenin kit (Boehringer Mannheim) and used for nonradioactive alkaline phosphatase detection.
RESULTS
The 32-kDA Polypeptide of Calf Thymus RNase HI Is Enzymatically Active.
Calf thymus RNase HI originally was purified to apparent homogeneity by Büsen (11, 25) and later by Rong and Carl (32). Those authors observed two protein bands of molecular mass 32 and 21 kDa, respectively, in SDS/PAGE gels of their highly purified preparations.
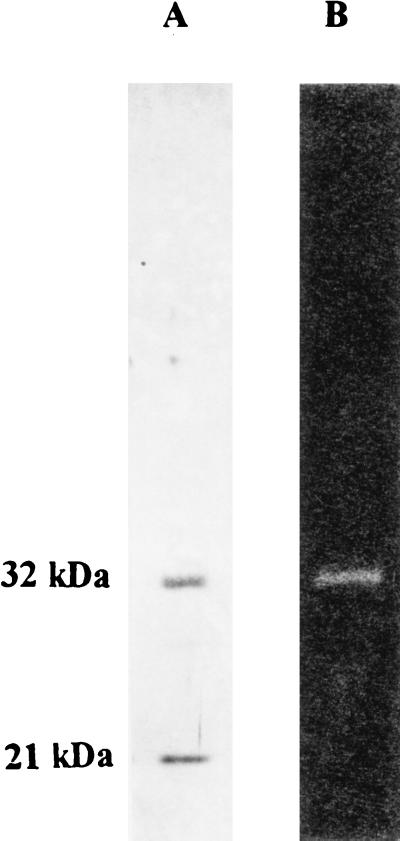
To get material for protein sequencing we purified calf thymus RNase HI according to the procedure of Büsen (25), using immunoaffinity chromatography as the final step. Our efforts starting from 120 g of calf thymus resulted in 280 μg of purified enzyme, which again displayed two bands of 32 kDa and 21 kDa, respectively, in SDS/PAGE gels stained with silver (Fig. 1A). When analyzing the preparation by renaturation gel assay (22), we found that the 32-kDA polypeptide exhibited RNase H activity (Fig. 1B). This finding is in accordance with the data of Rong and Carl (32) and allowed the conclusion that the 32-kDA polypeptide of our preparation carries the active center of the RNase HI. We therefore chose this polypeptide, of which it has, as yet, not been known whether it represented a subunit or a proteolytic breakdown product, for additional experiments.
Figure 1.
Analysis of immunoaffinity-purified calf thymus RNase HI. Samples containing 1 μg of protein each were subjected to SDS/PAGE (12% gels) and analyzed by silver staining (A) and tested for enzymatic activity by renaturation gel assay (B). Corresponding Mr positions are shown on the left.
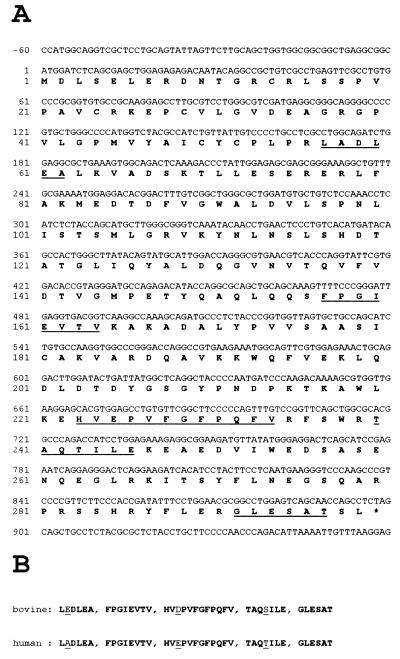
To isolate the 32-kDA polypeptide, the RNase H preparation was processed as described in Materials and Methods, including preparative SDS/PAGE, concentration on Vanderkerkhove gel, blotting onto nitrocellulose, and Coomassie brilliant blue R-250 staining. The stained spot was cut out and digested overnight with trypsin. HPLC purification of the tryptic peptides and Edman sequencing yielded five peptide sequences: LEDLEA, FPGIEVTV, HVDPVFGFPQFV, TAQSILE, and GLESAT.
Determination of the Sequence of a Human cDNA Found with the Aid of Peptides Originating from the 32-kDA Polypeptide of Bovine RNase HI.
By using the blast program and the EST database at the National Center for Biotechnology Information, we identified human ESTs coding for peptide motifs with identical or very similar sequences with respect to the bovine peptides described above (Fig. 2). Two of the five peptide sequences were identical between calf and human, each one of the other three displayed one amino acid substitution, among them two with similar amino acids. With the aid of these human ESTs it was possible to assemble an ORF potentially coding for a polypeptide of relative molecular mass 33,367. By using PCR techniques with different EST-derived primers and a human cDNA library from erythroleukemia cells, we were able to amplify, clone, and sequence the cDNA corresponding to the mRNA for a human protein. This cDNA encloses an ORF of 897 nt (299 aa), which defines a polypeptide with the expected molecular weight and a theoretical isoelectric point of 5.05. A computer search for known protein motifs, like RNA- or DNA-binding domains, did not yield conclusive results. Our finding demonstrates that the 33.4-kDa polypeptide is a subunit and not a breakdown product. Additionally, data (unpublished work) prove that the 21-kDa protein found in the calf thymus RNase HI preparation is also a distinct subunit and not a proteolytic degradation product of the bovine 32-kDA protein. Western blot experiments show that both proteins are immunologically distinct, and sequence analysis data of tryptic peptides of the 21-kDa protein did not fit to any part of the sequence of the human 33.4-kDa polypeptide, but allowed us to identify corresponding human ESTs, enabling us to determine the coding sequence of this ORF in the near future.
Figure 2.
(A) Sequence of cDNA and deduced amino acid sequence of the large subunit of human RNase HI. Peptide sequences used to identify the ORF are underlined. (B) Comparison of corresponding peptides originally obtained from purified calf thymus RNase HI, 32-kDA polypeptide, with those identified by analyses of human ESTs (EMBL accession no. AA262477 and GenBank accession no. H43540). Differences between the bovine peptides and the human homologues are underlined.
Sequence Comparison Uncovers a Relationship of Mammalian RNase HI to Prokaryotic RNase HII.
When searching the nonredundant protein database at the National Center for Biotechnology Information, we were surprised by finding highly significant homologies of the human 33.4-kDa RNase HI subunit (299 aa) with the experimentally identified RNase HII from E. coli (213 aa). Additionally, the recently published sequence of RNase HII from Staphylococcus pneumoniae (290 aa, see ref. 23) was detected. The authors of that work already provided evidence that bacterial RNase HII is very widely or universally distributed and has a homologous counterpart in human cells. Also numerous unidentified ORFs were discovered and unambiguously recognized as homologues of the large subunit of human RNase HI. Among these ORFs were those of the eukaryotes, Caenorhabditis elegans (1,254 aa), Schizosaccharomyces pombe (326 aa), and Saccharomyces cerevisiae (307 aa), as well as of three archaebacteria, Methanococcus jannaschii (230 aa), Archaeoglobus fulgidus (205 aa), and Methanobacterium thermoautotrophicum (206 aa), and of many eubacteria, like Bacillus subtilis (313 aa), Synechocystis sp. (190 aa), Mycobacterium tuberculosis (264 aa), Mycobacterium leprae (240 aa), Magnetospirillum sp. (201 aa), Haemophilus influenzae (197 aa), and Vibrio cholerae (162 aa).
An alignment of all of these sequences is shown in Fig. 3. By using the multiple sequence alignment with hierarchical clustering method of Corpet (33), five highly conserved motifs were detected in all 14 depicted sequences of the evolutionary divergent species. The motifs are: I, GxDEvGrGpxxGpxvxaa; II, gxxDSKxLsxxxrxxl; III, vxpxxvlxDgxxxp; IV, vxkgD/ExxxxxI/VsaASI/VxAkvxrdxxm; and V, gxxxxkGyptxxh (bold capital letters indicate identities and similarities, lowercase letters indicate 50% identities or more, and x is any amino acid). Another interesting feature is the existence of four insertions (marked A–D in Fig. 3) in the eukaryotic and archaebacterial sequences, which are absent in most of the eubacteria with B. subtilis and S. pneumonia as exceptions; in those two sequences only insertion A is absent. If one compares the sequences pairwise, the closest relative of the human RNase HI large subunit is the central part of a 1,254-aa hypothetical protein of C. elegans with a 45.5% overall identity (53.8% identity in a 251-aa overlap), followed by S. pombe (35.7% overall identity; 45.9% identity in a 231-aa overlap), S. cerevisiae (37, 5% overall identity; 42.8% identity in a 236-aa overlap), and the archaebacterium, M. jannaschii (24.4% overall identity; 30.7% in a 228-aa overlap). The eubacterium E. coli shows 24.7% overall identity with the human enzyme. Nevertheless the observed homology is significant.
Figure 3.
Multiple sequence alignment of the complete amino acid sequence of human RNase HI, large subunit, and related genes or corresponding ORFs (or parts thereof) from other eukaryotes, one archaebacterium, and a sample of eubacteria. The alignment was generated by using the multiple sequence alignment with hierarchical clustering method of Corpet (33), using sequences from H. sapiens (lanes 1, EMBL accession no. Z97029), C. elegans (lanes 2, EMBL Z66524), S. pombe (lanes 3, Swiss-Prot Q10236), S. cerevisiae (lanes 4, EMBL Z71348), M. jannaschii (lanes 5, PIR G64316), B. subtilis (lanes 6, EMBL Z75208), S. pneumoniae (lanes 7, GenBank U93576), Synechocystis sp. (lanes 8, EMBL D90899), M. tuberculosis (lanes 9, Swiss-Prot Q10793), M. leprae (lanes 10, EMBL Z97369), Magnetospirillum sp. (lanes 11, EMBL D32253), H. influenzae (lanes 12, Swiss-Prot P43808), E. coli (lanes 13, GenBank U70214), and V. cholerae (lanes 14, Swiss-Prot P52021). Numbering of amino acids above the alignment corresponds to the consecutive sequence of human RNase HI, large subunit (1–248 of 299 aa of this sequence are presented in the alignment, as only regions with existing consensus are shown). Identical or similar amino acids are marked in bold letters within the aligned sequences and in bold uppercase letters (identities) or bold asterisks (similarities) below the alignment to indicate the consensus (marked c). Levels of 50% identity and more within the aligned sequences are marked in lowercase letters. The following parameters were used for the alignment: symbol comparison table, blosum62; gap weight, 12; gap length weight, 2. Five regions of clustered identical and similar amino acids are indicated by roman numbers below the consensus. Four gaps occurring in the alignment mainly in eubacterial sequences are marked as A–D.
Expression of the 33.4-kDa RNase HI Subunit in E. coli.
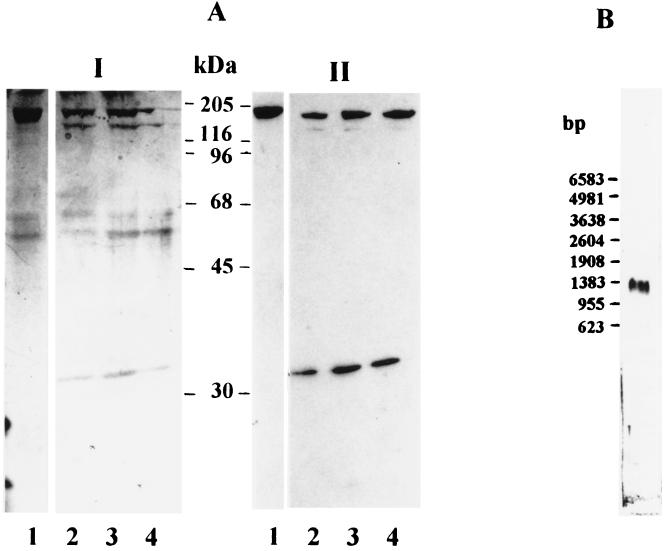
To confirm the identity of the sequenced ORF with mammalian RNase HI, we expressed it in E. coli as a fusion protein with β-galactosidase. By using PCR primers with appropriate cloning sites at both the 5′ and 3′ ends, we amplified the 33.4-kDa ORF and cloned it into the expression vector pXa1. Purification of the fusion protein and cleavage with the protease factor Xa led to a protein of around 33 kDa, a molecular mass that is in agreement with the expected size of 33.4 kDa. Fig. 4 shows the results of the experiments. As one can see in lanes 2–4 of Fig. 4AII, the protein is specifically recognized in Western blot analysis by the antiserum specific for calf thymus RNase HI (11), showing the close immunological relationship of the bovine and the human sequence.
Figure 4.
Expression of the large subunit of human RNase HI in E. coli and Northern blot analysis of total RNA from HEK-293 cells. (A) Purified large subunit-β-galactosidase fusion protein was digested with factor Xa protease for 0 min (lane 1), 220 min (lane 2), 240 min (lane 3), and 260 min (lane 4) and analyzed on a 12% gel. (I) Silver staining. (II) Western blotting with antiserum against calf thymus RNase HI (because of lack of a reaction product data of earlier time points are not shown). Positions of protein Mr markers are indicated. (B) Total RNA from HEK-293 cells was hybridized to digoxigenin-labeled probes specific for human RNase HI, large subunit. Positions of RNA Mr markers are shown on the left.
The gene coding for the large subunit of RNase HI is expressed in cultured HEK-293 cells as shown by the result of a Northern blot analysis depicted in Fig. 4B. Purified total RNA from HEK-293 cells was hybridized with a RNase HI-specific probe that detected a transcript corresponding to a length of around 1,200 nt.
DISCUSSION
Although mammalian RNase HI was the first RNase H to be identified (1), its primary structure is still unknown. Interest in this enzyme probably has been overwhelmed by the most illuminating work on RNase HI from the bacterium E. coli (17, 34), which is homologous to the RNase H domain of retroviral reverse transcriptases (35).
In the present work, we have reported the identification of the ORF coding for the large subunit of human RNase HI by using protein sequence information derived from the purified, enzymatically active bovine homologue. The obtained molecular mass of 33,367 for the human protein is in good agreement with the data of Eder and Walder (9) on the purified enzyme. To our surprise the deduced amino acid sequence does not contain any motifs exhibiting similarity to those typical and well established as being important for RNase H activity of the extensively studied enzymes, RNase HI of E. coli and the RNase H domain of reverse transcriptases. Rather, a highly significant similarity to E. coli RNase HII, an enzyme of which sequence parts responsible for activity are not yet known, was observed. Further sequence homology searches and alignments revealed that homologous ORFs were found in all completely sequenced genomes (with the exception of those from two mycoplasma species, which do not contain any gene homologous to known RNases H). Consideration of additional information from two partly sequenced genomes, those of the eukaryotes S. pombe and C. elegans, led to the identification of five common consensus domains (Fig. 3), which are potential candidates for substrate binding and enzymatic action. Interestingly, in most of the eubacterial sequences, if compared with the sequences from the archaebacterium and the eukaryotes, four small gaps, designated A–D in Fig. 3, are obvious. As the ORFs of B. subtilis and S. pneumoniae exhibit only gap A and parts of B and C a clear border between species with and without gaps (or insertions) cannot be drawn, and only more detailed studies concerning functionally important parts of the genes will reveal the significance of the observed differences.
Another distinction between the completely known genomes, however, may indeed be significant in evolutionary respects. Bacterial RNases, which were named RNases HI and which, as recently detected (36), are homologues of human RNase HII and S. cerevisiae RNase H1 (for better understanding, the relatives of the reverse transcriptase RNase H domains), are missing from the genomes of a number of organisms. So far these are the genomes of three archaebacteria (M. janaschii, A. fulgidus, and M. thermoautotrophicum; the latter two are not included in Fig. 3; for more information see http://www.ncbi.nlm.nih.gov/Entrez/Genome/org.html) and of the eubacteria, B. subtilis, Borrelia burgdorferi, and Aquifex aeolicus. In contrast, bacterial RNases HII occur in all genomes of free living prokaryotes that so far were completely sequenced. The fact that their eukaryotic counterparts, mammalian RNases HI and budding yeast RNase H(35), represent the main enzymes in the corresponding organisms, may underline the general importance of this type of RNase H for RNA–DNA hybrid metabolism. Eukaryotic RNase HII on the other side (corresponding to S. cerevisiae RNase H1 and eubacterial RNase HI) might have more specialized functions. Accordingly, as mentioned in the Introduction, the RNase HI of E. coli exhibits a regulatory role and is not essential for survival. We hope that by studying the RNase H equipment of further genomes the completion of which is expected for the near future we will be able to contribute to the ongoing debate about the evolutionary relationship between organisms, and especially to the question of the origin of eukaryotes, which recently has been raised again (37).
Acknowledgments
We thank Alexandra Bogusch for excellent technical assistance and Klaus Holzmann for helpful discussions concerning the computer analysis of ESTs. This research was supported by the Deutsche Forschungsgemeinschaft BU483/3 to W.B., by the Fonds zur Förderung der wissenschaftlichen Forschung in Österreich (Grant S 5806-MOB) to U.W., and the Anton Dreher Gedächtnisschenkung für medizinische Forschung (Grant 272/95) to P.F.
ABBREVIATIONS
- EST
expressed sequence tag
- HEK
human embryonal kidney
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. Z97029).
References
- 1. Stein H, Hausen P. Science. 1969;166:393–395. doi: 10.1126/science.166.3903.393. [DOI] [PubMed] [Google Scholar]
- 2.Henry C M, Ferdinand F-J, Knippers R. Biochem Biophys Res Commun. 1973;50:603–611. doi: 10.1016/0006-291x(73)91287-4. [DOI] [PubMed] [Google Scholar]
- 3.Miller H I, Riggs A D, Gill G N. J Biol Chem. 1973;248:2621–2624. [PubMed] [Google Scholar]
- 4.Tashiro F, Mita T, Higashinakagawa T. Eur J Biochem. 1976;65:123–130. doi: 10.1111/j.1432-1033.1976.tb10396.x. [DOI] [PubMed] [Google Scholar]
- 5.Wyers F, Huet J, Sentenac A, Fromageot P. Eur J Biochem. 1976;69:385–395. doi: 10.1111/j.1432-1033.1976.tb10372.x. [DOI] [PubMed] [Google Scholar]
- 6.Karwan R, Blutsch H, Wintersberger U. Biochemistry. 1983;22:5500–5502. [Google Scholar]
- 7.Moelling K, Bolognesi D P, Bauer H, Büsen W, Plassmann H W, Hausen P. Nat New Biol. 1971;234:240–243. doi: 10.1038/newbio234240a0. [DOI] [PubMed] [Google Scholar]
- 8.Büsen W, Peters J H, Hausen P. Eur J Biochem. 1977;74:203–208. doi: 10.1111/j.1432-1033.1977.tb11382.x. [DOI] [PubMed] [Google Scholar]
- 9.Eder P S, Walder J A. J Biol Chem. 1991;266:6472–6479. [PubMed] [Google Scholar]
- 10.Turchi J J, Huang L, Murante R S, Kim Y, Bambara R A. Proc Natl Acad Sci USA. 1994;91:9803–9807. doi: 10.1073/pnas.91.21.9803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Büsen W. J Biol Chem. 1980;255:9434–9443. [PubMed] [Google Scholar]
- 12.Masutani Ch, Enomoto T, Suzuki M, Hanaoka F, Ui M. J Biol Chem. 1990;265:10210–10216. [PubMed] [Google Scholar]
- 13.Kane C M. Biochemistry. 1988;27:3187–3193. doi: 10.1021/bi00409a010. [DOI] [PubMed] [Google Scholar]
- 14.Frank P, Albert S, Cazenave C, Toulmé J-J. Nucleic Acids Res. 1994;22:5247–5254. doi: 10.1093/nar/22.24.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giles R V, Ruddell C J, Spiller D G, Green J A, Tidd D M. Nucleic Acids Res. 1995;23:954–961. doi: 10.1093/nar/23.6.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies J F, Hostomska Z, Hostomsky Z, Jordan S R, Mathews D A. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 17.Katayanagi N, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Matsuzaki T, Morikawa K. Nature (London) 1990;347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 18.Wintersberger U, Wintersberger E. Trends Genet. 1987;3:198–202. [Google Scholar]
- 19.Kornberg A, Baker T A. DNA Replication. New York: Freeman; 1992. [Google Scholar]
- 20.Kogoma T. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itaya M. Proc Natl Acad Sci USA. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank P, Cazenave C, Albert S, Toulmé J-J. Biochem Biophys Res Commun. 1993;196:1552–1557. doi: 10.1006/bbrc.1993.2428. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y B, Ayalew S, Lacks S A. J Bacteriol. 1997;179:3828–3836. doi: 10.1128/jb.179.12.3828-3836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank P, Braunshofer-Reiter C, Wintersberger U. FEBS Lett. 1998;421:23–26. doi: 10.1016/s0014-5793(97)01528-7. [DOI] [PubMed] [Google Scholar]
- 25.Büsen W. J Biol Chem. 1982;257:7106–7108. [PubMed] [Google Scholar]
- 26.Vandekerckhove J, Rasmussen H H. In: Cell Biology: A Laboratory Handbook. Celis J E, editor. San Diego: Academic; 1994. pp. 359–368. [Google Scholar]
- 27.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Merril C R, Goldman D, VanKeuren M. Methods Enzymol. 1984;104:441–447. doi: 10.1016/s0076-6879(84)04111-2. [DOI] [PubMed] [Google Scholar]
- 30.Cazenave C, Frank P, Toulmé J-J, Büsen W. J Biol Chem. 1994;269:25185–25192. [PubMed] [Google Scholar]
- 31.Fernandez J, De Mott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 32.Rong Y W, Carl P L. Biochemistry. 1990;29:383–389. doi: 10.1021/bi00454a012. [DOI] [PubMed] [Google Scholar]
- 33.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kogoma T. J Bacteriol. 1994;176:1521–1523. doi: 10.1128/jb.176.5.1521-1523.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson M S, McClure M A, Feng D-F, Gray J, Doolittle R F. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank P., Braunshofer-Reiter, C., Poeltl, A. & Holzmann, K. (1998) Biol. Chem. in press. [DOI] [PubMed]
- 37.Martin W, Müller M. Nature (London) 1998;382:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]