Abstract
RecA is a 38-kDa protein from Escherichia coli that polymerizes on single-stranded DNA, forming a nucleoprotein filament that pairs with homologous duplex DNA and carries out strand exchange in vitro. To observe the effects of mismatches on the kinetics of the RecA-catalyzed recombination reaction, we used assays based upon fluorescence energy transfer that can differentiate between the pairing and strand displacement phases. Oligonucleotide sequences that produced 2–14% mismatches in the heteroduplex product of strand exchange were tested, as well as completely homologous and heterologous sequences. The equilibrium constant for pairing decreased as the number of mismatches increased, which appeared to result from both a decrease in the rate of formation and an increase in the rate of dissociation of the intermediates. In addition, the rate of strand displacement decreased with increasing numbers of mismatches, roughly in proportion to the number of mismatches. The equilibrium constant for pairing and the rate constant for strand displacement both decreased 6-fold as the heterology increased to 14%. These results suggest that discrimination of homology from heterology occurs during both pairing and strand exchange.
Keywords: recombination, energy transfer, kinetics, mismatches
RecA is a 38-kDa protein from Escherichia coli that is involved in recombination and repair (see ref. 1 for a current review). RecA has eukaryotic homologs from yeast to man that are also important in recombination and repair (2–5) and have similar biochemical activities to RecA (6–8). In vitro, RecA protein polymerizes on single-stranded DNA (ssDNA) in the presence of ATP, forming a right-handed nucleoprotein filament (9, 10). This filament can find and pair with homologous duplex, even when the target sequence is embedded in DNA with the complexity of the human genome (11). Once homologous alignment occurs, RecA protein carries out strand exchange (12, 13). The process of pairing and strand exchange is not stringent; strand exchange easily can bypass 3% difference in sequence between the ssDNA-RecA filament and duplex DNA even in the absence of ATP regeneration, and RecA protein can form D-loops between ssDNA and superhelical duplex DNA that differ in sequence by as much as 30% (14, 15). How does RecA distinguish homologous from heterologous sequences, and at which step, pairing or strand exchange? Previous studies have shown that mismatches affect the overall reaction and the rate of strand exchange promoted by RecA protein but have not succeeded in distinguishing the effects on pairing from the effects on strand exchange (16). In fact, only recently have assays been developed that kinetically can distinguish pairing from the early stages of strand exchange (17, 18).
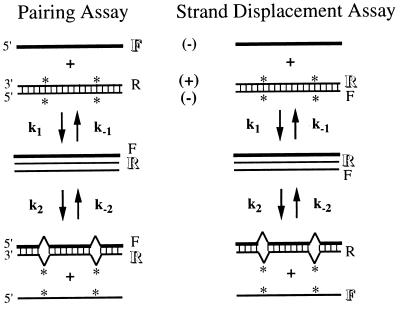
We developed assays based on energy transfer between fluorescent dyes on the ends of DNA oligonucleotides that can independently measure pairing and strand exchange in solution (18). In an assay designed to detect the initial pairing of the RecA-ssDNA filament with duplex DNA, fluorescein attached to the filament comes into proximity with rhodamine on the complementary strand of the duplex upon pairing, which quenches fluorescein because of energy transfer between the two dyes (Fig. 1; for a review of energy transfer, see ref. 19). By contrast, in the strand displacement assay, both dyes are located on the duplex, so that fluorescein initially is quenched and then increases in fluorescence as the two strands are separated during strand displacement (Fig. 1). The two assays allow the observation of pairing and strand displacement as separate phases with little apparent perturbation to the reaction. Oligonucleotide substrates provide a well defined system that is easily modified. In the experiments reported here, we used stopped-flow fluorescence spectroscopy to study reactions with mismatched substrates to determine whether the discrimination between homologous and heterologous sequences occurs in the pairing phase of the RecA reaction, during strand exchange, or both.
Figure 1.
Fluorescence assays for pairing and strand displacement. RecA-bound ssDNA, indicated by thicker lines, reacts with double-stranded DNA bearing complementary transversions, marked with ∗. Complementary strands are marked with (−) and (+). Intermediates form, which go on to strand exchange, creating mismatches, shown here as bulges. F represents fluorescein, R represents rhodamine, and letters in outline indicate which fluors are enhanced at a particular point in each assay. Symbols are assigned for the rate constants for each step of the reactions.
MATERIALS AND METHODS
Materials.
RecA protein was purified as described (20), with an additional step involving elution of the protein from a ssDNA-cellulose column with ATP (21). T4 polynucleotide kinase was supplied by New England Biolabs. ATP, phosphocreatine, and creatine phosphokinase were from Sigma. BSA was purchased from Boehringer Mannheim. DTT was provided by Promega. Proteinase K was from American Bioanalytical (Natick, MA). Fluorescent dyes were supplied by Molecular Probes. Primary amines with C6 linkers were from Glen Research (Sterling, VA).
Oligonucleotide Sequences.
M(−) oligonucleotide was 5′-TTG ATA AGA GGT CAT TTT TGC GGA TGG CTT AGA GCT TAA TTG CTG AAT CTG GTG CTG TAG CTC AAC ATG TTT TAA ATA TGC AA-3′. M(+) oligonucleotide (identical to M13 plus sequence beginning at nucleotide 182) was 5′-TTG CAT ATT TAA AAC ATG TTG AGC TAC AGC ACC AGA TTC AGC AAT TAA GCT CTA AGC CAT CCG CAA AAA TGA CCT CTT ATC AA-3′. Locations of transversions made in M(−) and M(+) sequences to produce substrates for mismatches are indicated by boldface type. M2(−) and M2(+) have changes at positions 24 and 60. M6(−) and M6(+) have changes at 12, 24, 36, 48, 60, and 72. M12(−) and M12(+) have transversions at all positions in bold. R83 is a random mixture of 83-mers with the same base composition as M(−). In the notation for fluorescent dyes, ⋅F after the oligonucleotide’s name indicates fluorescein is attached at the 3′ end, and ⋅R indicates rhodamine at the 5′ end.
Preparation of DNA Substrates.
Deoxyribo-oligonucleotide concentrations are given in terms of mols of molecules. Oligonucleotides were synthesized, labeled with 5-carboxyfluorescein or 5-carboxytetramethylrhodamine, and purified by denaturing polyacrylamide gel electrophoresis, as described (18, 22). Duplex oligonucleotides were prepared and checked by 5′ end-labeling with 32P (18, 23). All duplex substrates contained less than 5% single strands.
Standard Reaction Conditions.
Reactions were conducted at 37°C in buffer with 33 mM Pipes, pH 7.0/1 mM magnesium acetate/2 mM DTT/1.2 mM ATP/10 units of creatine phosphokinase per ml/10 mM phosphocreatine (16 mM was used in stopped-flow experiments). RecA protein filaments were formed on single-stranded oligonucleotides by incubation with one RecA monomer to three nucleotides for 2 min minimum. The M(−)⋅F or the R83⋅F oligonucleotide was used for the pairing assay, and M(−) or R83 with no fluorescein was used for the strand displacement assay (sequences listed above). The Mg2+ concentration was raised to 16 mM, and an equimolecular amount of duplex was added. Reactions were conducted with 0.06, 0.12, or 0.24 μM ssDNA in the RecA filament and the same amounts of duplex DNA. M(−)/M(+)⋅R duplex or permutations of that sequence [M2(−)/M2(+)⋅R, M6(−)/M6(+)⋅R, or M12(−)/M12(+)⋅R, sequences listed above] were used for the pairing assay, and M(−)⋅F/M(+)⋅R or its derivatives were used for the strand displacement assay.
Equilibrium Fluorescence Measurements.
Fluorescence emission was measured for the pairing and strand displacement assays on an SLM8000C spectrofluorometer (SLM–Aminco, Urbana, IL) (18). Measurements of fluorescein emission at 520 nm upon excitation at 493 nm were made before and after strand exchange. The change in fluorescence for each assay with each set of substrates was normalized by the change in fluorescence observed for the renaturation of M(−)⋅F-RecA filament and M(+)⋅R, M2(+)⋅R, M6(+)⋅R, or M12(+)⋅R (sequences listed above) with the same concentrations and conditions. The annealing reactions represented the maximum fluorescence change possible for both the pairing and strand displacement assays, assuming that the annealing reactions resulted in the formation of 100% duplex DNA. This assumption was confirmed by a gel electrophoresis assay for annealing with substrates that formed 0–6 mismatches (see below for details of assay, data not shown). Annealing with 12 mismatches produced 80% duplex DNA, which may reflect the instability of the heteroduplex after deproteinization, because the fluorescence of thermally renatured duplex oligonucleotide (which was 100% annealed, as indicated by gel assay) was only 5% lower than RecA-annealed duplex (data not shown). Enhancement of rhodamine emission by energy transfer was calculated by subtracting the background emission of fluorescein and rhodamine in the absence of energy transfer, as described (18).
Gel Electrophoresis Assay.
Both pairing and strand displacement assay substrates were 5′ end-labeled with 32P on the (−) strand of the duplex. The strand exchange reactions were conducted under standard conditions (see above), deproteinized, electrophoresed, and quantitated as described (18). In the gel assay for RecA-mediated annealing, 0.12 μM M(−)⋅F-RecA filament and 0.12 μM M(+)⋅R, M2(+)⋅R, M6(+)⋅R, or M12(+)⋅R were reacted under standard conditions for 5 min. To measure renaturation only in the presence of RecA protein, we added excess unlabeled M(−) oligonucleotide (1.2 μM, final concentration) before deproteinization and electrophoresis.
The reversibility of strand exchange was tested by reacting filament formed with 0.12 μM 32P-labeled M(−) and 6.67 μM RecA protein with 0.12 μM M(−)/M(+) or M6(−)/M6(+) duplex oligonucleotide. After 3 min, 0.12 μM unlabeled M(−) or M6(−) was added to drive the reaction back. Aliquots were taken before and 5 min after the addition of the unlabeled single strands. Aliquots were deproteinized and assayed by gel electrophoresis, as described above.
Stopped-Flow Spectrofluorometry.
The time courses of the pairing and strand displacement assays were observed at 520 nm upon excitation at 493 nm with a DX.17MV sequential stopped-flow spectrofluorometer (Applied Photophysics, Surrey, U.K.) (18). Final concentrations were 0.06, 0.12, or 0.24 μM ssDNA in the RecA filament and 0.06, 0.12, or 0.24 μM duplex oligonucleotide. The data were converted from fluorescence intensities to concentrations by normalizing the fluorescence time course at completion to the final concentration at equilibrium of the filament (in the pairing assay) or the displaced strand (in the strand displacement assay). The final concentrations were calculated by multiplying the total concentration by the percent completion of the reaction calculated previously by dividing the steady-state amplitude for the particular assay by the amplitude for annealing (Table 1) (18). Fluorescence amplitudes at 50 sec were used to normalize the assays with 12 mismatches because the reaction had not reached equilibrium.
Table 1.
Fluorescence amplitudes vs. yields at equilibrium
| Mismatches | Pairing assay, % reaction | Strand displ. assay, % reaction | Gel assay, % reaction |
|---|---|---|---|
| 0 | 62 | 36 | 44 |
| 2 | 54 | 31 | 34 |
| 6 | 48 | 22 | 24 |
| 12 | 31* | 19* | 12 |
| 83 | 4 | 0 | 0.5 |
Changes in fluorescein emission for the pairing and strand displacement assays (Fig. 1) are expressed as a percentage of the RecA-promoted annealing reaction between M(−)⋅F and M(+)⋅R or M(+)⋅R with mismatches. The values are the average of assays performed with 0.06, 0.12, or 0.24 μM ssDNA-RecA filament. Strand exchange yields with the fluorescent substrates were assayed by gel electrophoresis. Assays are described in Materials and Methods.
*Amplitudes at equilibrium (5 min) were larger than at 50 s (Fig. 2).
The data were fitted by using the program kinsim to generate theoretical curves based on the two-step model for strand exchange shown in Fig. 1 (24–26). The time courses for pairing and strand displacement were simultaneously simulated. Each of the four rate constants (k1, k−1, k2, and k−2) were independently varied until the theoretical curves for both the pairing and strand displacement assays could be superimposed on the experimental data for a particular concentration and number of mismatches. The initial rapid increase in fluorescence in the strand displacement assay indicated that intermediates also must be contributing to the increase in fluorescence (see ref. 18 for further discussion). The extent of this contribution cannot be determined experimentally, but we found that the assumption of a 10–20% increase in fluorescence upon the formation of intermediates fits the data. The assumption of values between 10% and 20% only changed k2, and by less than 8%.
The fitting procedure was repeated 8–14 times for each data set. The fits were of comparable quality: the sum of the residuals squared varied less than 5-fold. The mean values and standard deviations of rate constants were calculated (Table 3).
Table 3.
Rate constants for pairing and strand displacement
| [DNA] | Mismatches | k1(106M−1s−1) | k−1(10−2s−1) | Keq1(107M−1) | k2(10−2s−1) | k−2(106M−1s−1) |
|---|---|---|---|---|---|---|
| 0.24 μM | 0 | 0.98 ± 0.04 | 7.1 ± 0.2 | 1.38 ± 0.06 | 1.9 ± 0.1 | 0.43 ± 0.05 |
| 2 | 0.71 ± 0.03 | 9.3 ± 0.7 | 0.76 ± 0.04 | 2.1 ± 0.1 | 0.54 ± 0.05 | |
| 6 | 0.54 ± 0.03 | 8.6 ± 0.2 | 0.63 ± 0.04 | 0.88 ± 0.05 | 0.58 ± 0.05 | |
| 12 | 0.4 ± 0.1 | 14 ± 1 | 0.29 ± 0.07 | 0.30 ± 0.04 | 0.4 ± 0.1 | |
| 0.12 μM | 0 | 1.5 ± 0.2 | 6.0 ± 0.2 | 2.5 ± 0.2 | 1.9 ± 0.1 | 1.08 ± 0.08 |
| 2 | 1.27 ± 0.07 | 8.6 ± 0.5 | 1.48 ± 0.08 | 1.7 ± 0.1 | 1.1 ± 0.1 | |
| 6 | 0.97 ± 0.08 | 8.6 ± 0.8 | 1.13 ± 0.09 | 0.65 ± 0.07 | 0.9 ± 0.2 | |
| 12 | 0.65 ± 0.05 | 17 ± 2 | 0.38 ± 0.03 | 0.29 ± 0.02 | 0.5 ± 0.2 | |
| 0.06 μM | 0 | 1.9 ± 0.3 | 3.5 ± 0.3 | 3.1 ± 0.9 | 1.8 ± 0.3 | 1.75 ± 0.05 |
| 2 | 1.02 ± 0.04 | 4.7 ± 0.5 | 2.17 ± 0.09 | 2.1 ± 0.3 | 2.5 ± 0.5 | |
| 6 | 1.16 ± 0.08 | 4.1 ± 0.2 | 2.8 ± 0.2 | 0.62 ± 0.07 | 1.6 ± 0.5 | |
| Averages | 0 | 1.2 ± 0.1 | 6.6 ± 0.2 | 1.9 ± 0.1 | 1.9 ± 0.1 | 0.76 ± 0.07 |
| 0.12 μM | 2 | 0.99 ± 0.05 | 9.0 ± 0.6 | 1.12 ± 0.06 | 1.9 ± 0.1 | 0.82 ± 0.08 |
| & | 6 | 0.76 ± 0.06 | 8.6 ± 0.6 | 0.60 ± 0.07 | 0.77 ± 0.06 | 0.7 ± 0.1 |
| 0.24 μM | 12 | 0.53 ± 0.08 | 16 ± 2 | 0.34 ± 0.05 | 0.30 ± 0.03 | 0.5 ± 0.2 |
Rate constants were derived by fitting stopped-flow fluorescence data from the pairing and strand displacement assays according to the model in Fig. 1, as described in Materials and Methods.
RESULTS
Experimental Design.
In the experiments described here, we introduced mismatched bases into 83-mer oligonucleotides to determine at which stage RecA protein discriminates between homologous and nonhomologous sequences. Complementary transversions were made in the M(−)/M(+) duplex used for strand exchange, e.g., A to T in one strand, T to A in the other (sequences listed in Materials and Methods). Thus, the initial duplex was completely base-paired, but the heteroduplex product of strand exchange contained mismatches (Fig. 1). By substituting base pairs in the duplex oligonucleotide rather than the RecA filament strand, trivial causes of differences in efficiencies of reaction were avoided, such as changing the secondary structure of the single-stranded oligonucleotide. Because the substitutions were transversions, the base composition, and consequently the total number of hydrogen bonds, remained constant. A mixture of random 83-mers with the same percentage of each base as the M(−) oligonucleotide was used for the heterologous control.
Equilibrium Fluorescence Measurements.
To assess the effects of mismatches on the yields of pairing intermediates and products of strand exchange, we first made fluorescence measurements on the reactions at equilibrium. We chose to monitor changes in the emission of fluorescein in response to energy transfer because they are easier to observe than changes in the emission of rhodamine (18). However, we did perform controls in which we measured the sensitized emission of rhodamine to confirm energy transfer, as will be described below. We used two types of substrates in the fluorometric assays: pairing assay substrates, with fluorophores positioned for the detection of pairing (Fig. 1, Left), and strand displacement assay substrates, with fluorophores arranged to detect strand displacement (Fig. 1, Right). The amplitude, or the difference in fluorescein emission before and after strand exchange in each assay, was normalized by the amplitude of annealing as described in Materials and Methods, to reflect the fraction of substrates that was converted into intermediates and/or products.
The normalized amplitudes for the pairing and strand displacement assays were the same, within the error of the measurements, for a given set of substrates at the three concentrations of substrate that were systematically used (data not shown). The average values for the percent change in fluorescence relative to annealing are given in Table 1. The changes in fluorescence for both the pairing assay and the strand displacement assay decreased as the number of mismatches increased. Heterologous controls with a mixture of random 83-mers as the filament produced only 4% quenching of fluorescein in the pairing assay and no change in the strand displacement assay (Table 1). We confirmed that the changes in fluorescein emission observed during the course of the reaction were due to energy transfer, rather than to nonspecific effects, such as the distribution of RecA protein during the reaction, by measuring energy transfer to rhodamine (18). The changes in the sensitized emission of rhodamine decreased to the same extent as the changes in the quenched emission of fluorescein in both the pairing and strand displacement assays as the number of mismatches was increased (data not shown).
As shown in Fig. 1, the fluorometric assay for pairing detects both intermediates and heteroduplex products of the overall reaction. Consistent with that conclusion is the observation that the yields measured by the pairing assay were consistently higher than the yield of product measured by the strand displacement assay (Table 1). The difference in the yields between the pairing assay and the strand displacement assay should roughly equal the level of intermediates at equilibrium. A change in the yield of intermediates was not evident until 12 mismatches were introduced, and then the intermediates decreased by half (calculated from Table 1, column 2 − column 3). Similarly, the yield of products as assayed by the strand displacement assay also decreased by half relative to the fully homologous reaction when 12 mismatches were tested. This was the first indication that both the pairing step and the strand exchange step are affected by mismatches. The stopped-flow experiments below provided a more reliable and direct confirmation of these preliminary results.
Gel Assays for Strand Exchange.
We previously had observed good agreement between the measurement of strand displacement by the fluorometric assay and the measurement of strand exchange by the standard electrophoretic assay that requires deproteinization of the reaction mixture¶ (18). We performed similar experiments with all of the mismatched substrates studied here. We tested both pairing assay substrates and strand displacement assay substrates, which had the arrangements of fluorophores indicated in Fig. 1. We labeled the 5′ end of the minus strand of the duplex oligonucleotides with 32P to observe the displacement of the strand as a result of strand exchange. The differences in the positions of the fluorophores between the pairing assay substrates and the strand displacement assay substrates did not affect the yields of strand exchange (data not shown). Likewise, the percent of strand exchange was virtually the same at the three concentrations of substrates that were tested. The yields of strand exchange shown in Table 1 are the averages for both the pairing and strand displacement assay substrates at three concentrations. The yields of product observed by the gel assay correlate well with those measured by using the fluorometric strand displacement assay (Table 1).
The kinetic model used below to analyze the effects of mismatches on the phases of homologous pairing and strand exchange depends upon the assumption that the reaction involving mismatched substrates is reversible, as was demonstrated previously with fully homologous substrates (18). To test the reversibility of mismatched substrates, we compared strand exchange of substrates that produce six mismatches with those that are completely homologous. M(−) oligonucleotide labeled with 32P was used as the filament and was reacted with M(−)/M(+) or M6(−)/M6(+) duplex DNA. Strand exchange was initiated and allowed to come to equilibrium. Then, one stoichiometric equivalent of unlabeled ssDNA corresponding to the displaced strand, either M(−) or M6(−), was added to drive strand exchange back toward the reactants. Twice the normal stoichiometric amount of RecA protein was added initially, to ensure sufficient protein to coat the ssDNA that was added to drive the reaction in the reverse direction. As expected, the percentage of M(−)-32P incorporated in heteroduplex DNA during the forward reaction decreased as M(−)-32P was displaced by the unlabeled single strands that had been added to drive the reaction in the reverse direction (Table 2). The yields in the forward direction of strand exchange were higher than in previous experiments (Table 1), probably because twice as much RecA was present. However, in the reverse direction, when the ratio of RecA to ssDNA was the same as in previous experiments, the decrease in the percent of heteroduplex with M(−)-32P was consistent with the 44% yield of strand exchange observed previously (see reverse vs. predicted, Table 2; Table 1). The yield of the reverse reaction with mismatches was as good as the completely homologous reaction because mismatched heteroduplex was converted back to completely base-paired duplex DNA.
Table 2.
Reversibility of strand exchange
| Mismatches | Strand exchange, %
|
||
|---|---|---|---|
| Forward | Reverse | Predicted | |
| 0 | 69 | 39 | 39 |
| 6 | 58 | 34 | 32 |
After strand exchange in the forward direction leveled off, the reaction was driven in the reverse direction by the addition of unlabeled ssDNA corresponding to the displaced strand. The previously observed yield of 44% (Table 1, 0 mismatches) was used to predict the fraction of heteroduplex left after the reverse reaction. See Results for further details.
Stopped-Flow Kinetics.
The pairing and strand displacement assays were performed on a stopped-flow spectrofluorometer to obtain information on the kinetics of the reactions. The M(−)⋅F or M(−) oligonucleotide was preincubated with RecA protein and mixed in a 1:1 ratio with the appropriate duplex oligonucleotide that would generate 0–12 mismatches upon strand exchange. R83⋅F was reacted with M(−)/M(+)⋅R duplex as a heterologous control for pairing, which produced no detectable change in fluorescence, even at the highest concentrations of substrates used (data not shown). The amplitude of the pairing assay with 12 mismatches was so small at the lowest concentration tested that no change could be observed (data not shown). The fluorescence data were converted to concentrations as described in Materials and Methods and previously (18).
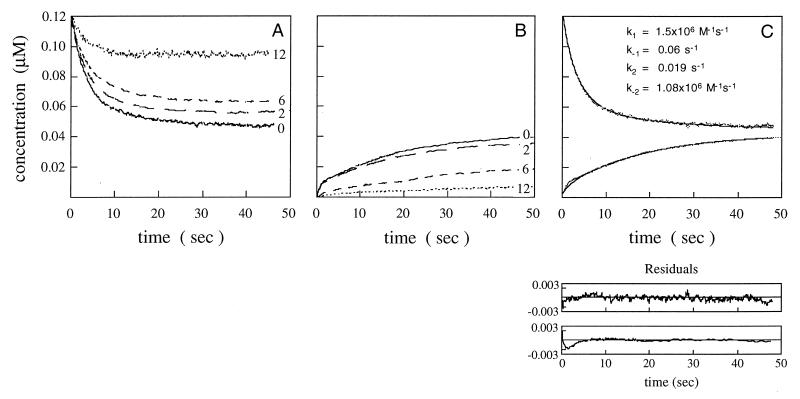
The change in rates with increasing numbers of mismatches can be seen even by inspection of the time courses of the assays (Fig. 2). The initial slope of the pairing assay curves, which reflects the rates of pairing, decreased from 0 mismatches to 12 (Fig. 2A). The amplitude of the pairing assay at early times (<10 sec) also decreased as the number of mismatches increased (Fig. 2A). This change in amplitudes was mainly a reflection of the equilibrium of pairing, because significant amounts of product had not formed yet (Fig. 2B). The strand displacement assay curves displayed even more clearly the decrease in rates and yields with the formation of mismatched products (Fig. 2B).
Figure 2.
Stopped-flow fluorescence assays for pairing and strand displacement. Filament (0.12 μM ssDNA 83-mer plus 3.33 μM RecA protein) and duplex 83-mer (0.12 μM) were reacted and fluorescein emission monitored at 520 nm upon excitation at 493 nm, as described in Materials and Methods. Fluorescent dyes were positioned as in Fig. 1. (A) Time courses for the pairing assay. The concentration is of the single-stranded oligonucleotide M(−)⋅F in the RecA filament. Solid line, 0 mismatches; long dashes, 2 mismatches; short dashes, 6 mismatches; dotted line, 12 mismatches. (B) Time courses for the strand displacement assay. Concentration is of the displaced strand (M(−)⋅F) plus 20% of the intermediate (see Materials and Methods for explanation). Same symbols as in A. (C) Simultaneous fitting of pairing and strand displacement data for 0 mismatches. Residuals of the fits are shown under the traces. (Upper) Pairing. (Lower) Strand displacement. (Inset) Values of rate constants used for the fit.
Inspection of the time courses suggested that mismatches affected both pairing and strand exchange, but kinetic analysis clearly was required to assess the separate effects on the phases of the reaction. To obtain rate constants, the data for the pairing assay and the strand displacement assay at a given concentration and number of mismatches were simultaneously fitted using theoretical curves generated by kinsim, based on the two-step model depicted in Fig. 1 (24–26). Because the experimental data did not provide enough constraints to determine the rate constants uniquely, the four rate constants had to be varied independently until the theoretical curves fit the experimental data. The initial slope of the pairing assay restrained k1, whereas the amplitude of the pairing assay limited the range of k−1. Similarly, k2 was defined by the slope and k−2 by the amplitude of the strand displacement assay. Fig. 2C shows one such fit with residuals to indicate the quality of the fits (see Materials and Methods for further details of the fitting procedure). The constants derived from the fitting procedure are listed in Table 3.
The most obvious change in rates was seen with k2, the rate constant for strand displacement. The rate constants were almost identical at different concentrations of substrates and seemed to decrease in proportion to the number of mismatches formed. The k2 values were almost the same for 0 and two mismatches, then decreased 3-fold as the number of mismatches increased from two to six, and another 2-fold as the mismatches increased from six to 12 (Table 3). The total change in k2 between 0 and 12 mismatches was 6-fold. No change in the rate constant for the back-reaction of strand displacement (k−2) was detected within the error of the measurements. If k−2 is constant, then the equilibrium constant for strand exchange, Keq2, must decrease 6-fold because Keq2 = k2/k−2. The absolute value of k2 in the reactions with no mismatches was smaller than the value determined in the previous paper, as was the amplitude for the strand displacement assay (Tables 1 and 2) (18). The reason for the change in efficiency is unknown, but the differences were consistent throughout this study.
The trends in the pairing rates were less striking than the changes in the strand exchange rates. The rate constant for pairing, k1, was consistently the largest for the substrates with no mismatches. In the reactions performed with 0.12 and 0.24 μM single-stranded oligonucleotide in the RecA filament, which had the highest signal-to-noise ratio, there seemed to be a consistent trend downward in k1 as the number of mismatches increased (Table 3). Conversely, k−1, the rate constant for the dissociation of pairing intermediates, increased as the number of mismatches increased. Combined, these two trends resulted in a 6-fold decrease in the equilibrium constant for pairing, Keq1, which is the ratio of k1/k−1.
Although the individual changes in the each of the rate constants for pairing were less than the change in rate constant for strand exchange, the additive effect on the equilibrium constant was the same. The introduction of 12 mismatches resulted in a 6-fold decrease in the equilibria for both pairing and strand exchange. From these data it appears that RecA exercises a similar degree of selection at both the pairing and the strand exchange steps of the RecA-catalyzed recombination reaction.
DISCUSSION
By using assays based on energy transfer, we have been able to assess separately the effects of heterology on the pairing and strand exchange phases of the RecA reaction. The rates of both phases are affected, indicating that detection of homology takes place during both. Incomplete homology results in the formation of fewer intermediates because of an apparent 2- to 3-fold reduction in the on-rate (k1) coupled with a similar increase in the off-rate (k−1) for pairing. The combined effect of these two trends is a 6-fold decrease in the equilibrium constant for pairing. The intermediates that survive the first selection process are subjected to a second screen in the form of strand exchange; k2 also decreases 6-fold as the number of mismatches increases from 0 to 12. Although the degree of selection that RecA protein exercises may seem small, these two screens are apparently sufficient for a RecA-bound oligonucleotide to find a homologous duplex target in human genomic DNA (11).
The rates obtained for strand displacement by computer simulation of the stopped-flow data were consistent among different concentrations and had low errors associated with them (Table 3). The errors in calculating the rate constants for pairing were larger, and prevented absolute conclusions about rates based solely on the analysis of rates by modeling. Some of the values were almost within the standard deviations of each other. However, the effects of mismatches on the rates of pairing were evident even from a qualitative look at the time courses of the pairing assay (Fig. 2A). As noted in Results, both the slope and the amplitude of the pairing assay at early times reflected the change in the rates of pairing. For example, the amplitude decreased as the number of mismatches increased, so Keq1 must have decreased, and either k1 or k−1 must have changed to account for the decrease in Keq1. Taken together, these observations and the analysis by computer simulation support the conclusion that the rates of pairing are changing in response to differences in sequence between the RecA filament and duplex DNA.
Strand exchange with completely homologous oligonucleotides is reversible (18, 27). In the present study, we demonstrated that strand exchange is reversible even with substrates that produce mismatches. Of course, reversibility was an important factor in the choice of kinetic models, but it also has implications for the fidelity of reactions promoted by RecA protein. If strand exchange were irreversible, then reactions with mismatches could reach 100% yield, if given enough time, despite decreases in k2. Instead, an equilibrium is established as unstable, mismatched heteroduplex products return to reactants. Reversibility reinforces the role of strand exchange as a second check for homology.
What are the mechanisms of discrimination between homology and heterology? The decrease in k2 is easily explained; mismatches present an energetic barrier, because branch migration is no longer isoenergetic. The increase in k−1 may be the result of the instability of intermediates that form between mismatched substrates. These two effects alone are enough to explain how RecA tests for homology. However, k1 appears to decrease as well, which suggests that mismatches present a barrier to the formation of intermediates. Perhaps in the search for homology initial contacts are made with a short segment of the filament, the effective “minimal recognition length,” which may be as short as eight bases (28, 29). If a mismatch is encountered within the minimal recognition length, then no intermediate is formed. The decrease in k1 may reflect the increase in the probability of encountering a mismatch within the minimal recognition length as the proportion of mismatches is increased.
According to our observations, RecA protein does not significantly discriminate between perfect and imperfect matches of sequence until the fraction of mismatches approaches 10%. However, in vivo, RecA is not called upon to promote strand exchange with absolute fidelity. The mismatch repair system, mut HLS, repairs mismatches and has been shown in vitro to halt RecA-catalyzed strand exchange between mismatched substrates (30–32). Mismatch repair adds a third, much more stringent screen for homology to the two imposed by RecA protein. Although a degree of fidelity is essential to the role of RecA protein, it instead has been adapted for the rapid and efficient elimination of ssDNA through recombination, whether the ssDNA is generated by damage to DNA or conjugation.
Acknowledgments
We thank Dr. M. Ibba and L. Lajoie for their help in maintaining the stopped-flow spectrofluorometer. We wish to express our appreciation to Dr. C. Freiden for providing the PC version of kinsim in advance of release, and Q. Dang for advice on its use. We are also thankful to Dr. D. Crothers for his helpful suggestions. We thank Z. Li for technical assistance and J. Zulkeski for secretarial assistance. This research was supported by National Institutes of Health Grant 5R37-GM33504-13.
ABBREVIATION
- ssDNA
single-stranded DNA
Footnotes
We use the term “strand displacement” to indicate the separation of the outgoing strand from the heteroduplex product, such that energy transfer can no longer occur. We are unable to specify if the separation is complete. “Strand exchange” designates the formation of products as assayed by gel electrophoresis. This measure is also ambiguous because the fate of intermediates after deproteinization is unknown.
References
- 1.Roca A I, Cox M M. Prog Nucleic Acid Res. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- 2.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 3.Bishop D K, Park D, Xu L, Kleckner N. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 4.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 5.Yoshimura Y, Morita T, Yamamoto A, Matsushiro A. Nucleic Acids Res. 1993;21:1665. doi: 10.1093/nar/21.7.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sung P. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 7.Baumann P, Benson F, West S. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R, Bazemore L R, Golub E I, Radding C M. Proc Natl Acad Sci USA. 1997;94:463–468. doi: 10.1073/pnas.94.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flory J, Radding C M. Cell. 1982;28:747–756. doi: 10.1016/0092-8674(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 10.Flory J, Tsang S S, Muniyappa K. Proc Natl Acad Sci USA. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrin L J, Camerini-Otero R D. Science. 1991;253:1494–1497. doi: 10.1126/science.1962209. [DOI] [PubMed] [Google Scholar]
- 12.DasGupta C, Shibata T, Cunningham R P, Radding C M. Cell. 1980;22:437–446. doi: 10.1016/0092-8674(80)90354-2. [DOI] [PubMed] [Google Scholar]
- 13.Cox M M, Lehman I R. Proc Natl Acad Sci USA. 1981;78:3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DasGupta C, Radding C M. Proc Natl Acad Sci USA. 1982;79:762–766. doi: 10.1073/pnas.79.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DasGupta C, Radding C M. Nature (London) 1982;295:71–73. doi: 10.1038/295071a0. [DOI] [PubMed] [Google Scholar]
- 16.Bianchi M E, Radding C M. Cell. 1983;35:511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- 17.Yancey-Wrona J E, Camerini-Otero R D. Curr Biol. 1995;5:1149–1158. doi: 10.1016/s0960-9822(95)00231-4. [DOI] [PubMed] [Google Scholar]
- 18.Bazemore L R, Takahashi M, Radding C M. J Biol Chem. 1997;272:14672–14682. doi: 10.1074/jbc.272.23.14672. [DOI] [PubMed] [Google Scholar]
- 19.Selvin P R. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 20.Shibata T, Cunningham R P, Radding C M. J Biol Chem. 1981;256:7557–7564. [PubMed] [Google Scholar]
- 21.Cox M M, McEntee K, Lehman I R. J Biol Chem. 1981;256:4676–4678. [PubMed] [Google Scholar]
- 22.Rao B J, Chiu S K, Radding C M. J Mol Biol. 1993;229:328–343. doi: 10.1006/jmbi.1993.1038. [DOI] [PubMed] [Google Scholar]
- 23.Chiu S K, Rao B J, Story R M, Radding C M. Biochemistry. 1993;32:13146–13155. doi: 10.1021/bi00211a025. [DOI] [PubMed] [Google Scholar]
- 24.Barshop B A, Wrenn R F, Freiden C. Anal Biochem. 1983;130:134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerle C T, Freiden C. Biochem J. 1989;258:381–387. doi: 10.1042/bj2580381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freiden C. Trends Biochem Sci. 1993;18:58–60. doi: 10.1016/0968-0004(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 27.Rosselli W, Stasiak A. J Mol Biol. 1990;216:335–352. doi: 10.1016/S0022-2836(05)80325-0. [DOI] [PubMed] [Google Scholar]
- 28.Thomas C A. Prog Nucleic Acid Res. 1966;5:315–337. doi: 10.1016/s0079-6603(08)60237-8. [DOI] [PubMed] [Google Scholar]
- 29.Hsieh P, Camerini-Otero C S, Camerini-Otero R D. Proc Natl Acad Sci USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modrich P, Lahue R. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 31.Worth J L, Clark S, Radman M, Modrich P. Proc Natl Acad Sci USA. 1994;91:3238–3241. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matic I, Rayssiguier C, Radman M. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]