Abstract
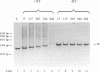
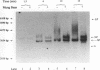
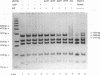
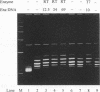
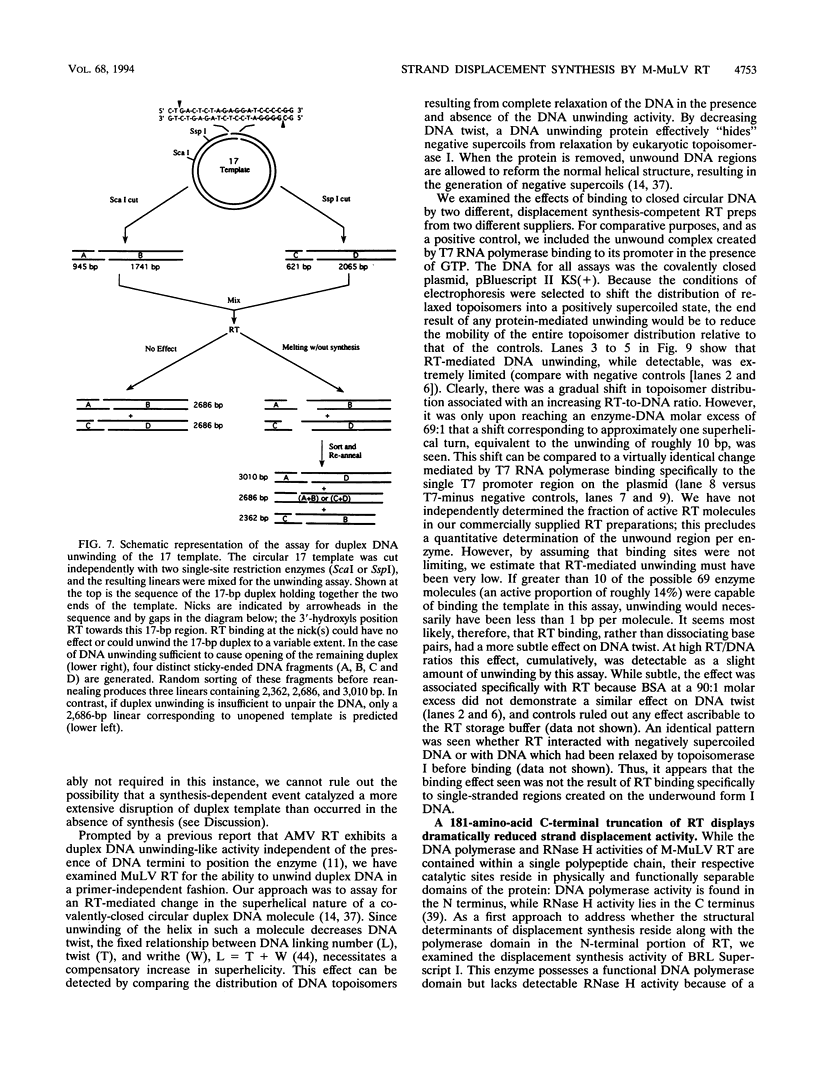
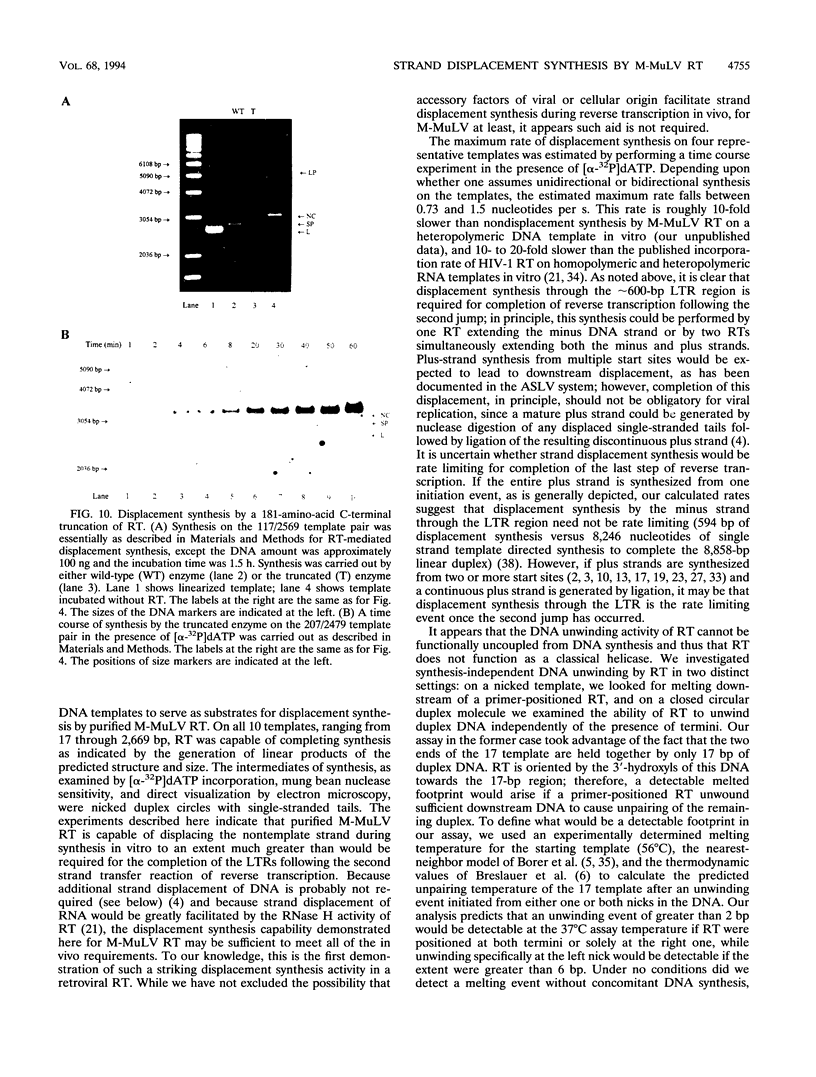
The accepted model of retroviral reverse transcription includes a circular DNA intermediate which requires strand displacement synthesis for linearization and creation of an integration-competent, long terminal repeat-flanked DNA product. We have used an in vitro model of this last step of reverse transcription to examine the role of the viral enzyme, reverse transcriptase (RT), in displacement synthesis. We show that Moloney murine leukemia virus RT possesses an activity which allows for displacement synthesis through a minimum of 1,334 bp of duplex DNA--an extent much greater than that required during in vivo reverse transcription and over 25-fold greater than has been previously demonstrated for a viral RT. RT does not function as a helicase in the classical sense but appears to closely couple duplex DNA melting with synthesis-driven translocation of the enzyme. In the absence of synthesis, the unwound region created by a primer-positioned RT appears to be no greater than 2 bp and does not advance along the template. Additionally, RT does not utilize ATP or any deoxynucleoside triphosphate not directly encoded by the template strand to catalyze processive duplex unwinding at a nick; nor does binding of the enzyme unwind duplex DNA in the absence of a 3' terminus. The approximate maximum chain elongation rate during strand displacement synthesis by Moloney murine leukemia virus RT falls between 0.73 and 1.5 nucleotides per s at 37 degrees C. The RNase H activity of RT does not appear to play a role in displacement synthesis; however, a 181-amino-acid C-terminal truncation of RT displays a dramatically reduced ability to catalyze synthesis through duplex DNA.
Full text
PDF
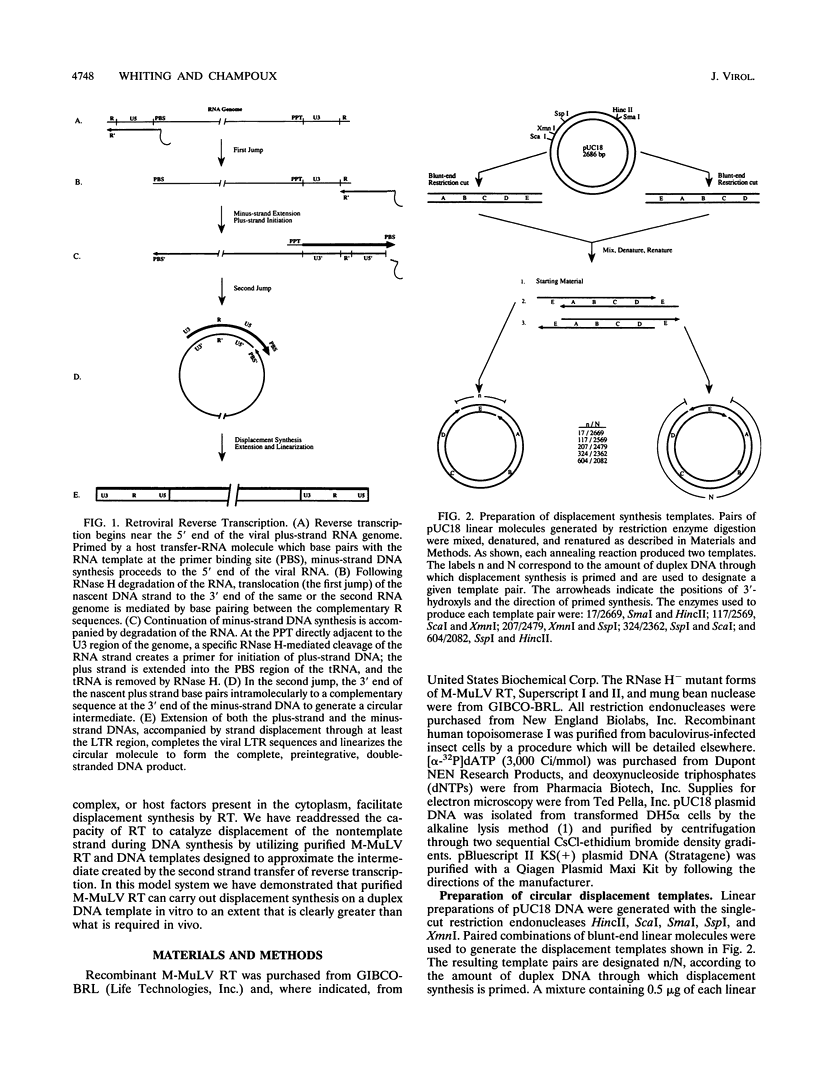

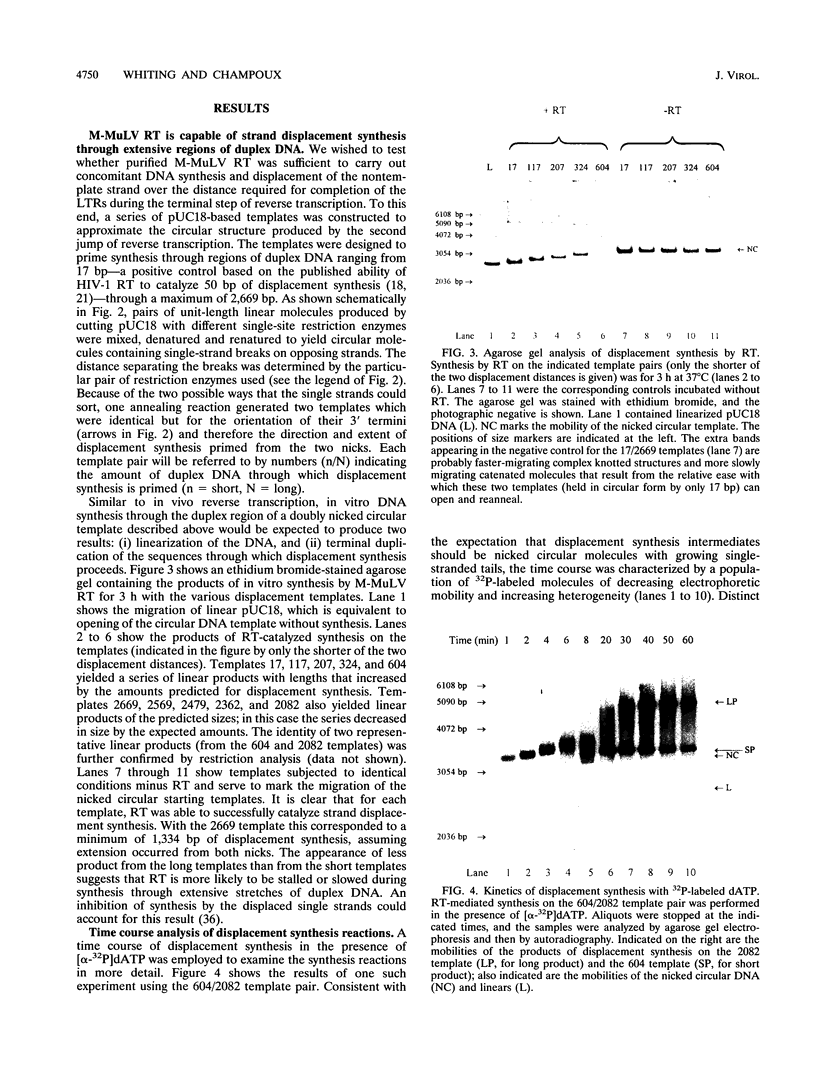

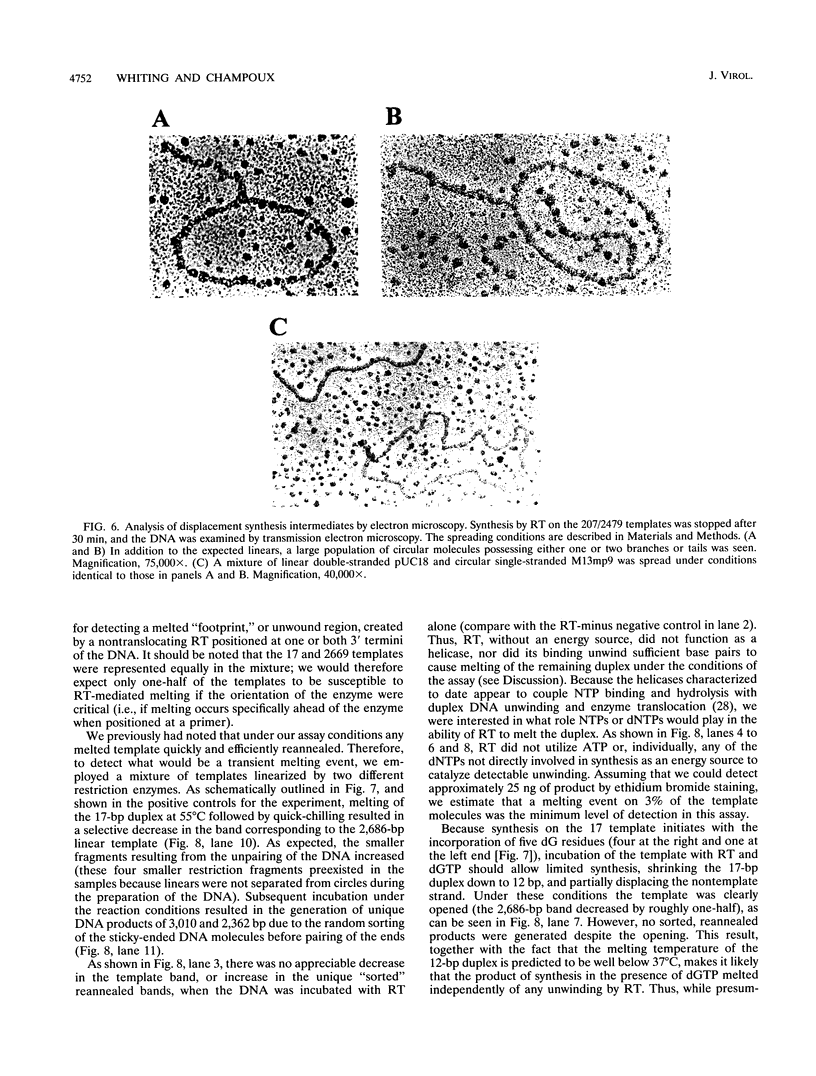




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blum H. E., Harris J. D., Ventura P., Walker D., Staskus K., Retzel E., Haase A. T. Synthesis in cell culture of the gapped linear duplex DNA of the slow virus visna. Virology. 1985 Apr 30;142(2):270–277. doi: 10.1016/0042-6822(85)90335-6. [DOI] [PubMed] [Google Scholar]
- Boone L. R., Skalka A. M. Viral DNA synthesized in vitro by avian retrovirus particles permeabilized with melittin. II. Evidence for a strand displacement mechanism in plus-strand synthesis. J Virol. 1981 Jan;37(1):117–126. doi: 10.1128/jvi.37.1.117-126.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer P. N., Dengler B., Tinoco I., Jr, Uhlenbeck O. C. Stability of ribonucleic acid double-stranded helices. J Mol Biol. 1974 Jul 15;86(4):843–853. doi: 10.1016/0022-2836(74)90357-x. [DOI] [PubMed] [Google Scholar]
- Breslauer K. J., Frank R., Blöcker H., Marky L. A. Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buiser R. G., DeStefano J. J., Mallaber L. M., Fay P. J., Bambara R. A. Requirements for the catalysis of strand transfer synthesis by retroviral DNA polymerases. J Biol Chem. 1991 Jul 15;266(20):13103–13109. [PubMed] [Google Scholar]
- Charneau P., Alizon M., Clavel F. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J Virol. 1992 May;66(5):2814–2820. doi: 10.1128/jvi.66.5.2814-2820.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneau P., Clavel F. A single-stranded gap in human immunodeficiency virus unintegrated linear DNA defined by a central copy of the polypurine tract. J Virol. 1991 May;65(5):2415–2421. doi: 10.1128/jvi.65.5.2415-2421.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collett M. S., Leis J. P., Smith M. S., Faras A. J. Unwinding-like activity associated with avian retrovirus RNA-directed DNA polymerase. J Virol. 1978 May;26(2):498–509. doi: 10.1128/jvi.26.2.498-509.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Rassart E., Zollinger M., Jolicoeur P. Synthesis of murine leukemia viral DNA in vitro: evidence for plus-strand DNA synthesis at both ends of the genome. J Virol. 1982 Apr;42(1):326–330. doi: 10.1128/jvi.42.1.326-330.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper H. B., Hearst J. E. Size of the unwound region of DNA in Escherichia coli RNA polymerase and calf thymus RNA polymerase II ternary complexes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):455–461. doi: 10.1101/sqb.1983.047.01.054. [DOI] [PubMed] [Google Scholar]
- Gilboa E., Mitra S. W., Goff S., Baltimore D. A detailed model of reverse transcription and tests of crucial aspects. Cell. 1979 Sep;18(1):93–100. doi: 10.1016/0092-8674(79)90357-x. [DOI] [PubMed] [Google Scholar]
- Hanas J. S., Bogenhagen D. F., Wu C. W. DNA unwinding ability of Xenopus transcription factor A. Nucleic Acids Res. 1984 Jan 25;12(2):1265–1276. doi: 10.1093/nar/12.2.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. D., Scott J. V., Traynor B., Brahic M., Stowring L., Ventura P., Haase A. T., Peluso R. Visna virus DNA: discovery of a novel gapped structure. Virology. 1981 Sep;113(2):573–583. doi: 10.1016/0042-6822(81)90185-9. [DOI] [PubMed] [Google Scholar]
- Hottiger M., Podust V. N., Thimmig R. L., McHenry C., Hübscher U. Strand displacement activity of the human immunodeficiency virus type 1 reverse transcriptase heterodimer and its individual subunits. J Biol Chem. 1994 Jan 14;269(2):986–991. [PubMed] [Google Scholar]
- Hsu T. W., Taylor J. M. Single-stranded regions on unintegrated avian retrovirus DNA. J Virol. 1982 Oct;44(1):47–53. doi: 10.1128/jvi.44.1.47-53.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. S., Temin H. M. Retroviral recombination and reverse transcription. Science. 1990 Nov 30;250(4985):1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- Huber H. E., McCoy J. M., Seehra J. S., Richardson C. C. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989 Mar 15;264(8):4669–4678. [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junghans R. P., Boone L. R., Skalka A. M. Products of reverse transcription in avian retrovirus analyzed by electron microscopy. J Virol. 1982 Aug;43(2):544–554. doi: 10.1128/jvi.43.2.544-554.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Kotewicz M. L., Sampson C. M., D'Alessio J. M., Gerard G. F. Isolation of cloned Moloney murine leukemia virus reverse transcriptase lacking ribonuclease H activity. Nucleic Acids Res. 1988 Jan 11;16(1):265–277. doi: 10.1093/nar/16.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec J. J., Tobaly-Tapiero J., Canivet M., Santillana-Hayat M., Flügel R. M., Périès J., Emanoil-Ravier R. Evidence for a gapped linear duplex DNA intermediate in the replicative cycle of human and simian spumaviruses. Nucleic Acids Res. 1988 Oct 25;16(20):9557–9565. doi: 10.1093/nar/16.20.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T. M. Helicase-catalyzed DNA unwinding. J Biol Chem. 1993 Feb 5;268(4):2269–2272. [PubMed] [Google Scholar]
- Lorch Y., Kornberg R. D. Near-zero linking difference upon transcription factor IID binding to promoter DNA. Mol Cell Biol. 1993 Mar;13(3):1872–1875. doi: 10.1128/mcb.13.3.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson S. W., Fay P. J., Bambara R. A. Mechanism of inhibition of the avian myeloblastosis virus deoxyribonucleic acid polymerase by adriamycin. Biochemistry. 1980 May 13;19(10):2089–2096. doi: 10.1021/bi00551a014. [DOI] [PubMed] [Google Scholar]
- Metzger W., Hermann T., Schatz O., Le Grice S. F., Heumann H. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template.primer complexes. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):5909–5913. doi: 10.1073/pnas.90.13.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Rattray A. J., Champoux J. J. The role of Moloney murine leukemia virus RNase H activity in the formation of plus-strand primers. J Virol. 1987 Sep;61(9):2843–2851. doi: 10.1128/jvi.61.9.2843-2851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon J. E. Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry. 1992 May 12;31(18):4473–4479. doi: 10.1021/bi00133a013. [DOI] [PubMed] [Google Scholar]
- Rychlik W., Spencer W. J., Rhoads R. E. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990 Nov 11;18(21):6409–6412. doi: 10.1093/nar/18.21.6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samadashwily G. M., Dayn A., Mirkin S. M. Suicidal nucleotide sequences for DNA polymerization. EMBO J. 1993 Dec 15;12(13):4975–4983. doi: 10.1002/j.1460-2075.1993.tb06191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucier J. M., Wang J. C. Angular alteration of the DNA helix by E. coli RNA polymerase. Nat New Biol. 1972 Oct 11;239(93):167–170. doi: 10.1038/newbio239167a0. [DOI] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Cywinski A., Smith J. K. Discontinuities in the DNA synthesized by an avian retrovirus. J Virol. 1983 Dec;48(3):654–659. doi: 10.1128/jvi.48.3.654-659.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telesnitsky A., Goff S. P. RNase H domain mutations affect the interaction between Moloney murine leukemia virus reverse transcriptase and its primer-template. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1276–1280. doi: 10.1073/pnas.90.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobaly-Tapiero J., Kupiec J. J., Santillana-Hayat M., Canivet M., Peries J., Emanoil-Ravier R. Further characterization of the gapped DNA intermediates of human spumavirus: evidence for a dual initiation of plus-strand DNA synthesis. J Gen Virol. 1991 Mar;72(Pt 3):605–608. doi: 10.1099/0022-1317-72-3-605. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]