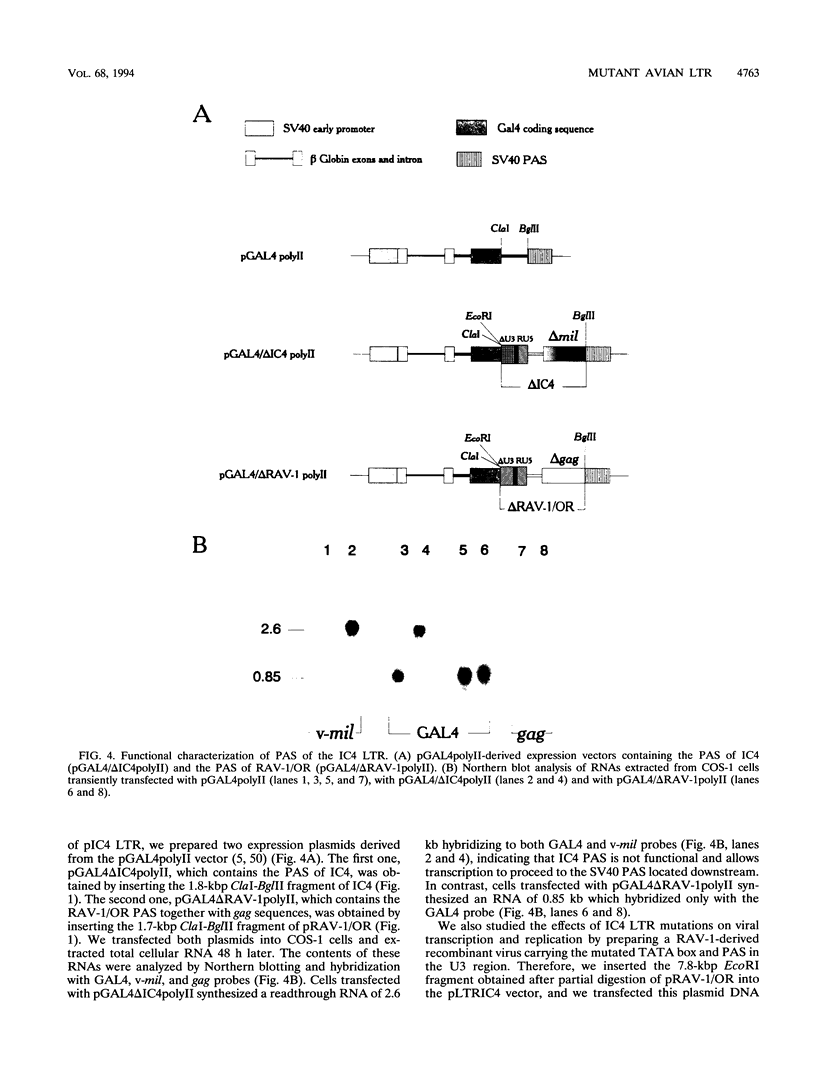
Abstract
We previously reported that infection of chicken embryonic neuroretina cells with Rous-associated virus type 1 leads to the frequent occurrence of spliced readthrough transcripts containing viral and cellular sequences. Generation of such chimeric transcripts constitutes a very early step in oncogene transduction. We report, here, the isolation of a c-mil transducing retrovirus, designated IC4, which contains a highly mutated U3 sequence in which 48% of A is converted to G. Functional analysis of this variant U3 indicated that these mutations do not impair viral transcription and replication; however, they abolish functioning of its polyadenylation signal, thus allowing readthrough transcription of downstream cellular sequences. On the basis of these results, we designed a nonreplicative retroviral vector, pIC4Neo, expressing the neomycin resistance (Neo(r)) gene under the control of the IC4 long terminal repeat. Infection of nondividing neuroretina cells with virus produced by a packaging cell line transfected with pIC4Neo occasionally resulted in sustained cell proliferation. Two independent G418-resistant proliferating cultures were found to express hybrid RNAs containing viral and cellular sequences. These sequences were characterized by reverse transcription-PCR and were identified in both cultures, suggesting that proliferation was correlated with a common integration locus. These results indicate that IC4Neo virus functions as a useful insertional mutagen and may allow identification of genes potentially involved in regulation of cell division.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bass B. L., Weintraub H., Cattaneo R., Billeter M. A. Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell. 1989 Feb 10;56(3):331–331. doi: 10.1016/0092-8674(89)90234-1. [DOI] [PubMed] [Google Scholar]
- Benchaibi M., Mallet F., Thoraval P., Savatier P., Xiao J. H., Verdier G., Samarut J., Nigon V. Avian retroviral vectors derived from avian defective leukemia virus: role of the translational context of the inserted gene on efficiency of the vectors. Virology. 1989 Mar;169(1):15–26. doi: 10.1016/0042-6822(89)90036-6. [DOI] [PubMed] [Google Scholar]
- Bizub D., Katz R. A., Skalka A. M. Nucleotide sequence of noncoding regions in Rous-associated virus-2: comparisons delineate conserved regions important in replication and oncogenesis. J Virol. 1984 Feb;49(2):557–565. doi: 10.1128/jvi.49.2.557-565.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulukos K. E., Pognonec P., Begue A., Galibert F., Gesquière J. C., Stéhelin D., Ghysdael J. Identification in chickens of an evolutionarily conserved cellular ets-2 gene (c-ets-2) encoding nuclear proteins related to the products of the c-ets proto-oncogene. EMBO J. 1988 Mar;7(3):697–705. doi: 10.1002/j.1460-2075.1988.tb02865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Harris B. A. Plasmids for the cloning and expression of full-length double-stranded cDNAs under control of the SV40 early or late gene promoter. Nucleic Acids Res. 1983 Oct 25;11(20):7119–7136. doi: 10.1093/nar/11.20.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cosset F. L., Ronfort C., Molina R. M., Flamant F., Drynda A., Benchaibi M., Valsesia S., Nigon V. M., Verdier G. Packaging cells for avian leukosis virus-based vectors with various host ranges. J Virol. 1992 Sep;66(9):5671–5676. doi: 10.1128/jvi.66.9.5671-5676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R., Raymond K., Ju G. Transcriptional activity of avian retroviral long terminal repeats directly correlates with enhancer activity. J Virol. 1985 Feb;53(2):515–521. doi: 10.1128/jvi.53.2.515-521.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix J. L. Control of Rous sarcoma virus RNA translation and packaging by the 5' and 3' untranslated sequences. J Mol Biol. 1986 Jun 5;189(3):421–434. doi: 10.1016/0022-2836(86)90314-1. [DOI] [PubMed] [Google Scholar]
- Eychène A., Béchade C., Marx M., Laugier D., Dezélée P., Calothy G. Molecular and biological properties of c-mil transducing retroviruses generated during passage of Rous-associated virus type 1 in chicken neuroretina cells. J Virol. 1990 Jan;64(1):231–238. doi: 10.1128/jvi.64.1.231-238.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder M. P., Eychène A., Barnier J. V., Calogeraki I., Calothy G., Marx M. Common mechanism of retrovirus activation and transduction of c-mil and c-Rmil in chicken neuroretina cells infected with Rous-associated virus type 1. J Virol. 1991 Jul;65(7):3633–3640. doi: 10.1128/jvi.65.7.3633-3640.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felder M. P., Laugier D., Eychene A., Calothy G., Marx M. Occurrence of alternatively spliced leader-delta onc-poly(A) transcripts in chicken neuroretina cells infected with Rous-associated virus type 1: implication in transduction of the c-mil/c-raf and c-Rmil/B-raf oncogenes. J Virol. 1993 Nov;67(11):6853–6856. doi: 10.1128/jvi.67.11.6853-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G. M., Parsons J. T. Identification of transcriptional elements within the long terminal repeat of Rous sarcoma virus. Mol Cell Biol. 1983 Oct;3(10):1834–1845. doi: 10.1128/mcb.3.10.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Merlino G. T., Willingham M. C., Pastan I., Howard B. H. The Rous sarcoma virus long terminal repeat is a strong promoter when introduced into a variety of eukaryotic cells by DNA-mediated transfection. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6777–6781. doi: 10.1073/pnas.79.22.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda S., Rao A. S., Kim Y. W., Guntaka R. V. Identification of sequences in the long terminal repeat of avian sarcoma virus required for efficient transcription. Virology. 1988 Jan;162(1):243–247. doi: 10.1016/0042-6822(88)90415-1. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herman S. A., Coffin J. M. Differential transcription from the long terminal repeats of integrated avian leukosis virus DNA. J Virol. 1986 Nov;60(2):497–505. doi: 10.1128/jvi.60.2.497-505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz R. A., Skalka A. M. Generation of diversity in retroviruses. Annu Rev Genet. 1990;24:409–445. doi: 10.1146/annurev.ge.24.120190.002205. [DOI] [PubMed] [Google Scholar]
- Kimelman D., Kirschner M. W. An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell. 1989 Nov 17;59(4):687–696. doi: 10.1016/0092-8674(89)90015-9. [DOI] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Tsichlis P., Khoury G. Multiple enhancer domains in the 3' terminus of the Prague strain of Rous sarcoma virus. Nucleic Acids Res. 1984 Aug 24;12(16):6427–6442. doi: 10.1093/nar/12.16.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Luciw P. A., Bishop J. M., Varmus H. E., Capecchi M. R. Location and function of retroviral and SV40 sequences that enhance biochemical transformation after microinjection of DNA. Cell. 1983 Jul;33(3):705–716. doi: 10.1016/0092-8674(83)90013-2. [DOI] [PubMed] [Google Scholar]
- Luo G. X., Chao M., Hsieh S. Y., Sureau C., Nishikura K., Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990 Mar;64(3):1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx M., Crisanti P., Eychène A., Béchade C., Laugier D., Ghysdaël J., Pessac B., Calothy G. Activation and transduction of c-mil sequences in chicken neuroretina cells induced to proliferate by infection with avian lymphomatosis virus. J Virol. 1988 Dec;62(12):4627–4633. doi: 10.1128/jvi.62.12.4627-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx M., Eychène A., Laugier D., Béchade C., Crisanti P., Dezélée P., Pessac B., Calothy G. A novel oncogene related to c-mil is transduced in chicken neuroretina cells induced to proliferate by infection with an avian lymphomatosis virus. EMBO J. 1988 Nov;7(11):3369–3373. doi: 10.1002/j.1460-2075.1988.tb03209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W., Maroney P. A., Goodwin R. G., Rottman F. M., Crittenden L. B., Raines M. A., Kung H. J. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell. 1985 Jul;41(3):719–726. doi: 10.1016/s0092-8674(85)80052-0. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Yoo C., Kim U., Murray J. M., Estes P. A., Cash F. E., Liebhaber S. A. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991 Nov;10(11):3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara P. J., Nichol S. T., Horodyski F. M., Holland J. J. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell. 1984 Apr;36(4):915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- Pathak V. K., Temin H. M. Broad spectrum of in vivo forward mutations, hypermutations, and mutational hotspots in a retroviral shuttle vector after a single replication cycle: substitutions, frameshifts, and hypermutations. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6019–6023. doi: 10.1073/pnas.87.16.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessac B., Calothy G. Transformation of chick embryo neuroretinal cells by Rous sarcoma virus in vitro: induction of cell proliferation. Science. 1974 Aug;185(4152):709–710. doi: 10.1126/science.185.4152.709. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rosson D., Dugan D., Reddy E. P. Aberrant splicing events that are induced by proviral integration: implications for myb oncogene activation. Proc Natl Acad Sci U S A. 1987 May;84(10):3171–3175. doi: 10.1073/pnas.84.10.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda P., García-Barreno B., Melero J. A. Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations). Virology. 1994 Feb;198(2):653–662. doi: 10.1006/viro.1994.1077. [DOI] [PubMed] [Google Scholar]
- Sanger F. Determination of nucleotide sequences in DNA. Science. 1981 Dec 11;214(4526):1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- Sealey L., Chalkley R. At least two nuclear proteins bind specifically to the Rous sarcoma virus long terminal repeat enhancer. Mol Cell Biol. 1987 Feb;7(2):787–798. doi: 10.1128/mcb.7.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M. C., Neckameyer W. S., Hayward W. S., Smith R. E. Genetic determinants of neoplastic diseases induced by a subgroup F avian leukosis virus. J Virol. 1987 Apr;61(4):1203–1212. doi: 10.1128/jvi.61.4.1203-1212.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Vennstrom B., Hayman M. J., Enrietto P. J. Nucleotide sequence of HBI, a novel recombinant MC29 derivative with altered pathogenic properties. J Virol. 1985 Dec;56(3):969–977. doi: 10.1128/jvi.56.3.969-977.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stein B., Rahmsdorf H. J., Steffen A., Litfin M., Herrlich P. UV-induced DNA damage is an intermediate step in UV-induced expression of human immunodeficiency virus type 1, collagenase, c-fos, and metallothionein. Mol Cell Biol. 1989 Nov;9(11):5169–5181. doi: 10.1128/mcb.9.11.5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Influence of sequences in the long terminal repeat and flanking cell DNA on polyadenylation of retroviral transcripts. J Virol. 1993 Oct;67(10):6265–6269. doi: 10.1128/jvi.67.10.6265-6269.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Mechanism of transduction by retroviruses. Science. 1992 Feb 14;255(5046):841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- Swain A., Coffin J. M. Polyadenylation at correct sites in genome RNA is not required for retrovirus replication or genome encapsidation. J Virol. 1989 Aug;63(8):3301–3306. doi: 10.1128/jvi.63.8.3301-3306.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. W., Smith J. E., Cooperman B. S., Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. W., Yoo C., Wrabetz L., Kamholz J., Buchhalter J., Hassan N. F., Khalili K., Kim S. U., Perussia B., McMorris F. A. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol Cell Biol. 1990 Oct;10(10):5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]