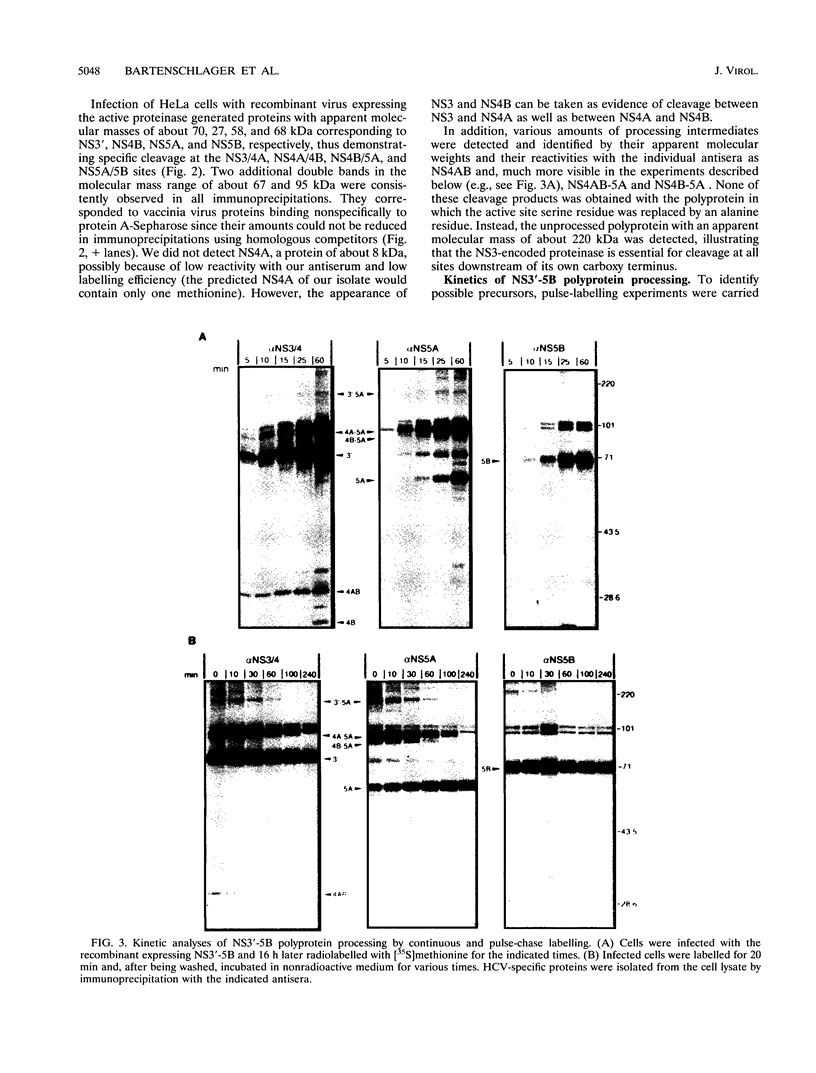
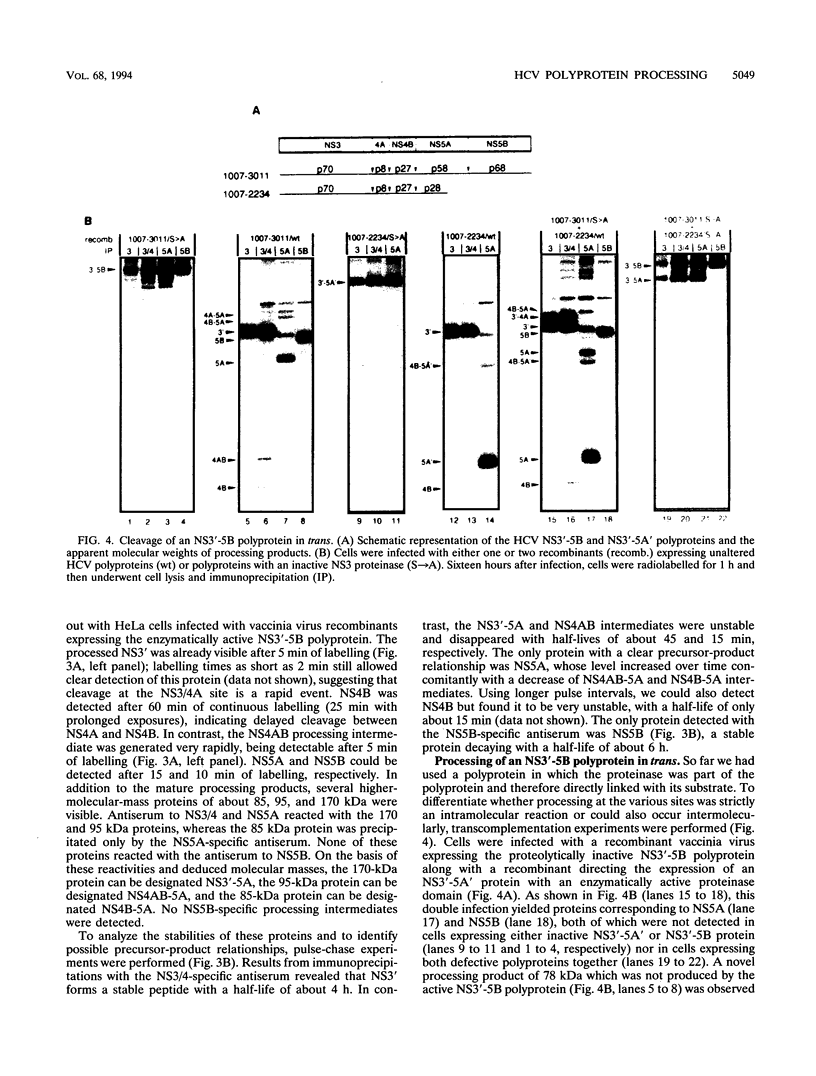
Abstract
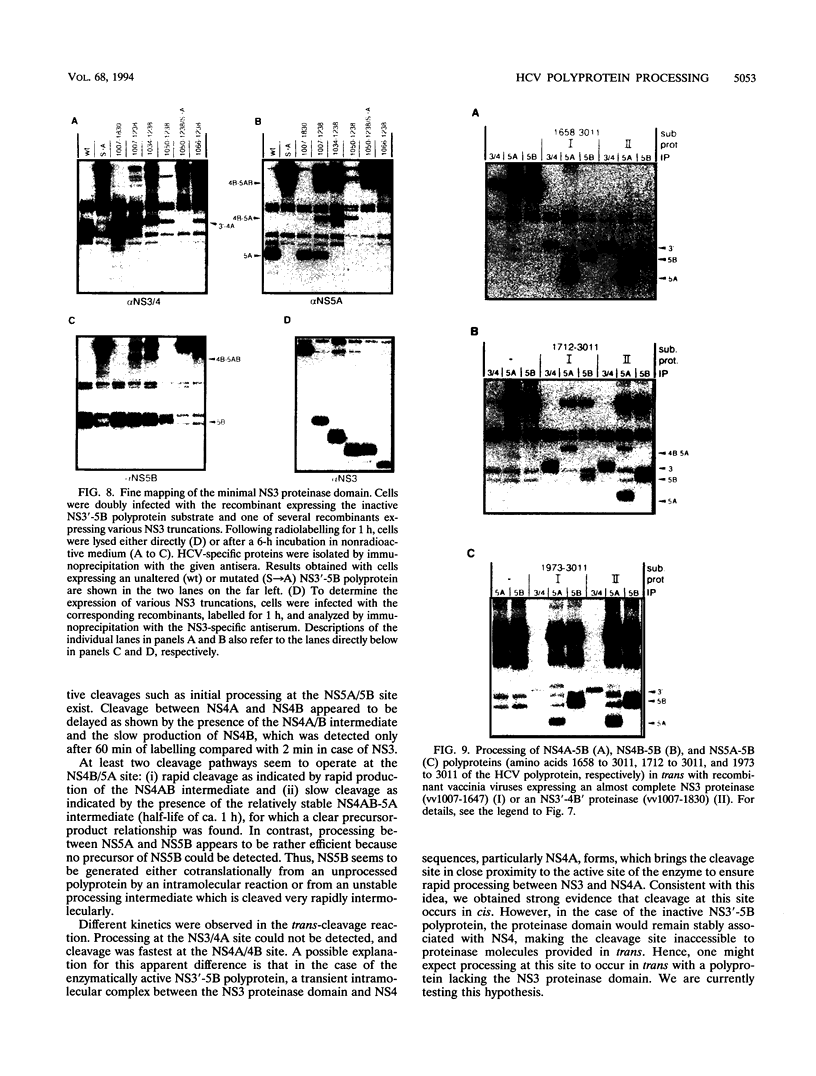
Recombinant vaccinia viruses were used to study the processing of hepatitis C virus (HCV) nonstructural polyprotein precursor. HCV-specific proteins and cleavage products were identified by size and by immunoprecipitation with region-specific antisera. A polyprotein beginning with 20 amino acids derived from the carboxy terminus of NS2 and ending with the NS5B stop codon (amino acids 1007 to 3011) was cleaved at the NS3/4A, NS4A/4B, NS4B/5A, and NS5A/5B sites, whereas a polyprotein in which the putative active site serine residue was replaced by an alanine remained unprocessed, demonstrating that the NS3-encoded serine-type proteinase is essential for cleavage at these sites. Processing of the NS3'-5B polyprotein was complex and occurred rapidly. Discrete polypeptide species corresponding to various processing intermediates were detected. With the exception of NS4AB-5A/NS5A, no clear precursor-product relationships were detected. Using double infection of cells with vaccinia virus recombinants expressing either a proteolytically inactive NS3'-5B polyprotein or an active NS3 proteinase, we found that cleavage at the NS4A/4B, NS4B/5A, and NS5A/5B sites could be mediated in trans. Absence of trans cleavage at the NS3/4A junction together with the finding that processing at this site was insensitive to dilution of the enzyme suggested that cleavage at this site is an intramolecular reaction. The trans-cleavage assay was also used to show that (i) the first 211 amino acids of NS3 were sufficient for processing at all trans sites and (ii) small deletions from the amino terminus of NS3 selectively affected cleavage at the NS4B/5A site, whereas more extensive deletions also decreased processing efficiencies at the other sites. Using a series of amino-terminally truncated substrate polyproteins in the trans-cleavage assay, we found that NS4A is essential for cleavage at the NS4B/5A site and that processing at this site could be restored by NS4A provided in cis (i.e., together with the substrate) or in trans (i.e., together with the proteinase). These results suggest that in addition to the NS3 proteinase, NS4A sequences play an important role in HCV polyprotein processing.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arias C. F., Preugschat F., Strauss J. H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993 Apr;193(2):888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R., Ahlborn-Laake L., Mous J., Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993 Jul;67(7):3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R., Kuhn C., Schaller H. Expression of the P-protein of the human hepatitis B virus in a vaccinia virus system and detection of the nucleocapsid-associated P-gene product by radiolabelling at newly introduced phosphorylation sites. Nucleic Acids Res. 1992 Jan 25;20(2):195–202. doi: 10.1093/nar/20.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukh J., Purcell R. H., Miller R. H. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8234–8238. doi: 10.1073/pnas.90.17.8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahour A., Falgout B., Lai C. J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992 Mar;66(3):1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Grakoui A., Rice C. M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991 Nov;65(11):6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers T. J., Hahn C. S., Galler R., Rice C. M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- Chambers T. J., Weir R. C., Grakoui A., McCourt D. W., Bazan J. F., Fletterick R. J., Rice C. M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien D. Y., Choo Q. L., Tabrizi A., Kuo C., McFarland J., Berger K., Lee C., Shuster J. R., Nguyen T., Moyer D. L. Diagnosis of hepatitis C virus (HCV) infection using an immunodominant chimeric polyprotein to capture circulating antibodies: reevaluation of the role of HCV in liver disease. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10011–10015. doi: 10.1073/pnas.89.21.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989 Apr 21;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Collett M. S. Molecular genetics of pestiviruses. Comp Immunol Microbiol Infect Dis. 1992 Jul;15(3):145–154. doi: 10.1016/0147-9571(92)90087-8. [DOI] [PubMed] [Google Scholar]
- Colombo M., Kuo G., Choo Q. L., Donato M. F., Del Ninno E., Tommasini M. A., Dioguardi N., Houghton M. Prevalence of antibodies to hepatitis C virus in Italian patients with hepatocellular carcinoma. Lancet. 1989 Oct 28;2(8670):1006–1008. doi: 10.1016/s0140-6736(89)91016-7. [DOI] [PubMed] [Google Scholar]
- Eckart M. R., Selby M., Masiarz F., Lee C., Berger K., Crawford K., Kuo C., Kuo G., Houghton M., Choo Q. L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993 Apr 30;192(2):399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- Falgout B., Pethel M., Zhang Y. M., Lai C. J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991 May;65(5):2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., McCourt D. W., Wychowski C., Feinstone S. M., Rice C. M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993 May;67(5):2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada S., Watanabe Y., Takeuchi K., Suzuki T., Katayama T., Takebe Y., Saito I., Miyamura T. Expression of processed core protein of hepatitis C virus in mammalian cells. J Virol. 1991 Jun;65(6):3015–3021. doi: 10.1128/jvi.65.6.3015-3021.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Kato N., Ootsuyama Y., Nakagawa M., Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Akagi T., Mori S., Kakiuchi N., Kato N., Tanaka T., Kimura K., Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993 Aug;67(8):4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata M., Mizushima H., Tanji Y., Komoda Y., Hirowatari Y., Akagi T., Kato N., Kimura K., Shimotohno K. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10773–10777. doi: 10.1073/pnas.90.22.10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton M., Weiner A., Han J., Kuo G., Choo Q. L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991 Aug;14(2):381–388. [PubMed] [Google Scholar]
- Hsu H. H., Donets M., Greenberg H. B., Feinstone S. M. Characterization of hepatitis C virus structural proteins with a recombinant baculovirus expression system. Hepatology. 1993 May;17(5):763–771. [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Harada S., Suzuki R., Watanabe Y., Inoue Y., Saito I., Miyamura T. Expression of processed envelope protein of hepatitis C virus in mammalian and insect cells. J Virol. 1992 Mar;66(3):1425–1431. doi: 10.1128/jvi.66.3.1425-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. H., Purcell R. H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preugschat F., Yao C. W., Strauss J. H. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990 Sep;64(9):4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito I., Miyamura T., Ohbayashi A., Harada H., Katayama T., Kikuchi S., Watanabe Y., Koi S., Onji M., Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby M. J., Choo Q. L., Berger K., Kuo G., Glazer E., Eckart M., Lee C., Chien D., Kuo C., Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993 Jun;74(Pt 6):1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Holmes E. C., Cha T. A., Chan S. W., McOmish F., Irvine B., Beall E., Yap P. L., Kolberg J., Urdea M. S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993 Nov;74(Pt 11):2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- Spaete R. R., Alexander D., Rugroden M. E., Choo Q. L., Berger K., Crawford K., Kuo C., Leng S., Lee C., Ralston R. Characterization of the hepatitis C virus E2/NS1 gene product expressed in mammalian cells. Virology. 1992 Jun;188(2):819–830. doi: 10.1016/0042-6822(92)90537-y. [DOI] [PubMed] [Google Scholar]
- Stål P., Hultcrantz R. Iron increases ethanol toxicity in rat liver. J Hepatol. 1993 Jan;17(1):108–115. doi: 10.1016/s0168-8278(05)80530-6. [DOI] [PubMed] [Google Scholar]
- Suzich J. A., Tamura J. K., Palmer-Hill F., Warrener P., Grakoui A., Rice C. M., Feinstone S. M., Collett M. S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993 Oct;67(10):6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Hirohata T., Koga S., Sugimachi K., Kanematsu T., Ohryohji F., Nawata H., Ishibashi H., Maeda Y., Kiyokawa H. Hepatitis C and hepatitis B in the etiology of hepatocellular carcinoma in the Japanese population. Cancer Res. 1991 Jun 1;51(11):2842–2847. [PubMed] [Google Scholar]
- Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993 Jul;67(7):4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskerchen M., Collett M. S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991 Sep;184(1):341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]