Abstract
A key strategy to achieve regulated gene expression in higher eukaryotes is to prevent illegitimate signal-independent activation by imposing robust control on the dismissal of corepressors. Here, we report that many signaling pathways, including Notch, NFkB, and nuclear receptor ligands, are subjected to a dual repression “check point” based on distinct corepressor complexes. Gene activation requires the release of both CtBP1/2- and NCoR/SMRT-dependent repression, through the coordinate action of two highly related exchange factors, the transducer β-like proteins TBL1 and TBLR1, that license ubiquitylation and degradation of CtBP1/2 and NCoR/SMRT, respectively. Intriguingly, their function and differential specificity resides in only five specific Ser/Thr phosphorylation site differences, regulated by direct phosphorylation at the level of the promoter, as exemplified by the role of PKCδ in TBLR1-dependent dismissal of NCoR. Thus, our data reveal a strategy of dual- factors repression checkpoints, in which dedicated exchange factors serve as sensors for signal- specific dismissal of distinct corepressors, with specificity imposed by upstream signaling pathways.
Keywords: TBL1, TBLR1, nuclear receptors, N-CoR, CtBP, ubiquitylation, repression, transcription
The mechanisms that control the precisely- regulated switch from gene repression to gene activation represent a central question in transcriptional regulation. Activating stimuli induce recruitment of specific DNA-binding transcription factors to their gene target promoters, mediate cytoplasmic-nuclear translocation of transcription factors and cofactors, and promote the local reorganization of the transcriptional machinery recruited by bifunctional transcription factors, favoring the exchange between corepressor and coactivators complexes. This last regulatory event being mediated by posttranslational and conformational changes of the DNA-bound transcription factor as well as by dedicated dismissal of the corepressors via translocation and/or degradation (Kleine-Kohlbrecher et al., 2006; Li et al., 2003; Perissi and Rosenfeld, 2005; Privalsky, 2004). The corepressors N-CoR and SMRT, initially identified as nuclear receptor corepressors, have also been implicated, in association with several histone deacetylase (HDACs) proteins, in repressive events mediated by a variety of unrelated transcription factors, which regulate diverse cellular processes in development and homeostasis (Asahara et al., 1999; Lee et al., 2000; Wong and Privalsky, 1998; Xu et al., 1998).
Transcriptional activation mediated by liganded nuclear receptors and by other regulated transcription factors is characterized by dismissal of corepressors, including the NCoR/SMRT corepressor complex, and the recruitment of a series of coactivator complexes harboring specific enzymatic activities (Heinzel et al., 1997; Metivier et al., 2003; Privalsky, 2004; Rosenfeld et al., 2006). TBL1 and TBLR1, two highly related F-box/WD-40 containing factors, are intrinsic components of the corepressor machinery, being initially identified as components of an N-CoR corepressor complex (Guenther et al., 2001; Li et al., 2000; Tomita et al., 2004; Yoon et al., 2003; Zhang et al., 2002a). Intriguingly, they are also important for activation by acting as specific adaptors for the recruitment of the ubiquitin conjugating/19S proteasome complex that mediate the exchange of corepressors for coactivators (Perissi et al., 2004). Despite of their high homology, there is some evidence that TBL1 and TBLR1 show distinct functions, as only TBLR1 appears to be required for activation by AP1 and RAR. In addition, in contrast to TBLR1, TBL1 is required for activation even in absence of the NCoR/SMRT/HDAC3 corepressor complex, suggesting that it may be responsible for the ubiquitylation and the dismissal of a different class of independently recruited corepressors (Perissi et al., 2004). Indeed, recent reports have suggested that multiple repressor complexes may be used combinatorially and be recruited by nuclear receptors in a sequential fashion, similarly to what described for numerous coactivator complexes (An et al., 2004; Liu and Bagchi, 2004; Metivier et al., 2003; Perissi and Rosenfeld, 2005; Yoon and Wong, 2006). Among the others, the corepressor CtBP (E1A C-terminal binding protein) was originally identified as a binding partner of the viral oncoprotein E1A and later reported to be an integral component of several corepressor complexes, with its repressive activities being partly explained by its NAD-dependent dehydrogenase activity (Chinnadurai, 2002; Kumar et al., 2002; Shi et al., 2003; Zhang et al., 2002b). Interestingly, for both NCoR/SMRT and CtBP corepressors examples have been reported of phosphorylation-dependent redistribution of cellular localization and relief of their repressive functions, often in association with the corepressor turnover and degradation. This includes the phosphorylation of SMRT Ser2410 by IKKα, important for the derepression of NFκB and Notch target genes (Fernandez-Majada et al., 2007; Hoberg et al., 2004); the phosphorylation of CtBP by Pak1, regulating its cellular localization and repression activities (Barnes et al., 2003); and the modulation of NCoR in response to IL1β inflammatory stimuli or to AKT activation during astrocytes differentiation (Baek et al., 2002; Hermanson et al., 2002). However, the functional relevance of the cooperation between different corepressors and the molecular mechanisms used to control the dismissal of each player upon signal stimulation are still unclear.
Here, we report an unexpected interaction between TBL1 and CtBP, and describe how both TBL1 and TBLR1 are required by a large number of regulated transcription factors to simultaneously overcome the repressive functions of the CtBP1/2 and NCoR/SMRT corepressor complexes, respectively, with TBL1 being specifically required to mediate ubiquitylation and degradation of CtBP. In addition, we describe that the functions and the specificity of these two highly- related exchange factors is tightly regulated by signal-induced phosphorylation events at the level of target gene promoters, as exemplified by the role of TBLR1 phosphorylation at Ser 123 by PKCδ upon retinoic acid or estrogen stimulation.
TBL1- and TBLR1-dependent dismissal of different corepressors is required for activation
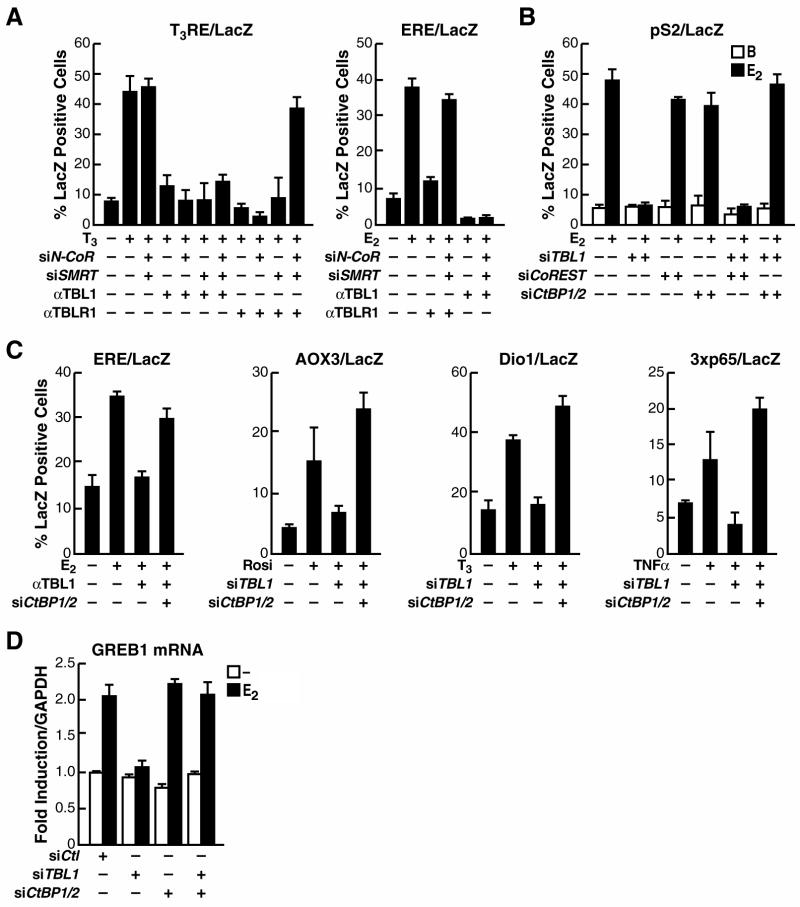
TBL1 and TBLR1 have proved to be widely required for transcriptional activation by a series of transcription factors, including ER, AR, TRβ, PPARγ and NFκB (Perissi et al., 2004). For all of them TBLR1 served as a dedicated factor for the ubiquitylation and dismissal of NCoR and SMRT; however, TBL1 was found to be required for signal-dependent transcriptional activation even in the absence of the NCoR/SMRT corepressors (Fig. 1A) (Perissi et al., 2004), implying that another class of repressive molecule must require TBL1 for its regulated dismissal. To investigate its potential role in promoting the dismissal of such an additional repressor complex, recruited in a NCoR-independent way, we screened for corepressors that, on removal, would be sufficient to rescue ligand-dependent activation in the absence of TBL1 in a single cell siRNA microinjection assay, using transcriptional activation of a LacZ reporter driven by the estrogen-responsive pS2 promoter in ER-positive MCF7 cells as readout. We found that the corepressors CtBP1 and CtBP2 proved to have this function, as the actions of TBL1 was no longer required in the absence of CtBP1/2 (Fig. 1B). Downregulation of other NCoR-independent corepressors, for example COREST, did not affect the requirement for TBL1 (Fig 1B and data not shown). To extend the significance of this finding, we investigated whether CtBP1/2 removal was sufficient to rescue activation for all the other transcription factors previously determined to require TBL1 (Perissi et al., 2004), including an ERE response element in response to estrogen, the PPARγRE-containing AOX3 promoter in response to Rosiglitazone, the TRβRE-containing Dio1 promoter in response to TRIAC, and a 3x-p65 response element in response to TNFα (Fig. 1C). In both Hela and MCF7 cells, transcriptional activation of all these transcription units required TBL1, but could be rescued if CtBP1/2 were downregulated by specific siRNA microinjection (Fig. 1C and data not shown). Finally, we also investigated transcriptional activation of the endogenous estrogen-target gene GREB1 in MCF7 cells, finding that the requirement for TBL1 in estrogen dependent induction could again be rescued by downregulation of CtBP1/2 (Fig 1D). Together these data revealed that TBL1 was required for transcriptional activation only in the presence of CtBP1/2, suggesting that TBL1 exerts a dedicated role in promoting the dismissal of the CtBP corepressor complex, analogous to the role of TBLR1 in mediating dismissal of the NCoR/SMRT complex.
Figure 1. TBL1 and TBLR1 are required for dismissal of distinct corepressors.
(A) Single-cell nuclear microinjection of purified IgGs against either TBL1 or TBLR1 inhibited transcriptional activation of a T3RE-dependent and of an ERE-dependent LacZ reporter in Rat1 cells. Depletion of TBLR1, but not TBL1, could be rescued by downregulating NCoR and SMRT by specific siRNAs. (B) A LacZ reporter driven by a 1.2kb fragment of the pS2 promoter was used in MCF7 to screen for other corepressors removal by siRNAs. Downregulation of CtBP1and CtBP2 fully rescued estrogen-induced activation in absence of TBL1. (C) αTBL1 IgGs microinjection blocked activation of LacZ reporters driven by an ERE response element, a PPARγ-responsive fragment of the AOX3 promoter, a TR-responsive fragment of the Dio1 promoter and a 3xp65 response element. All were rescued by removal of CtBP1 and CtBP2 by specific siRNAs microinjection. (D) Analysis of GREB1 mRNA expression in transiently transfected MCF7 cells by RT-PCR at 4h after induction. Expression of the GAPDH gene was used for normalization. Validation of the siRNA used here is shown in Supplemental Figure S1.
Distinct corepressors occupy regulated target genes
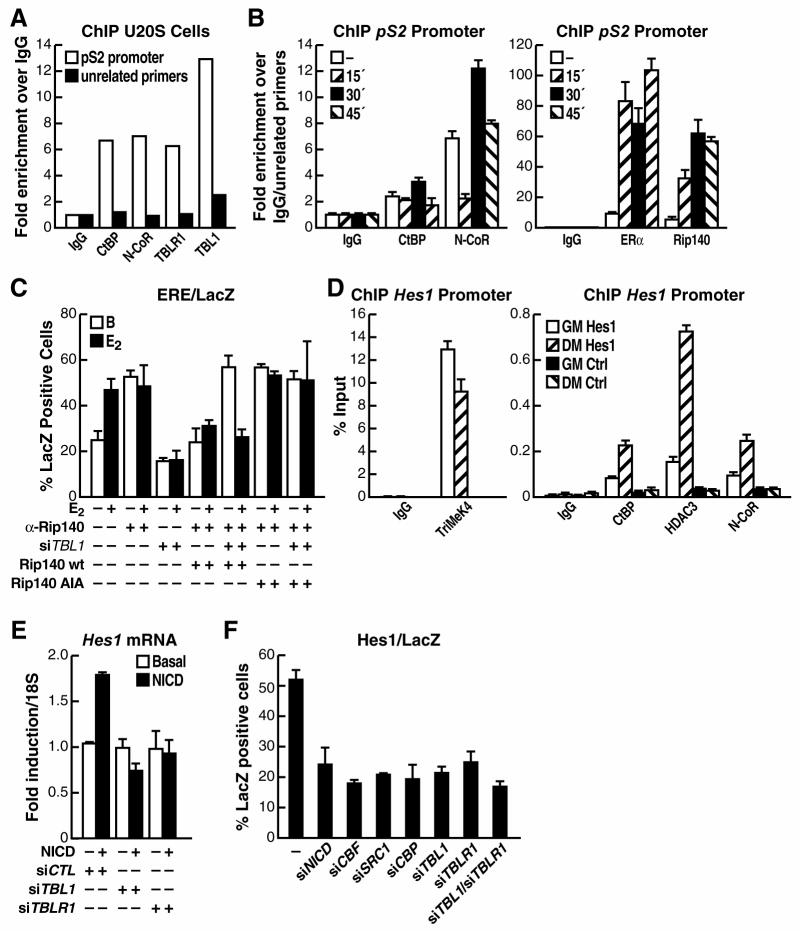
Interestingly, these observations implied that two distinct corepressor complexes would be required for regulation of gene transcription by many classes of transcription factors. We found that, indeed both CtBP and NCoR/SMRT corepressors were physically present on an estrogen regulated transcription unit, pS2 (Masiakowski et al., 1982), in either the full repressed state in ER-negative U2OS cells or upon activation at the end of the first productive cycle in estrogen responsive MCF7 cells (Fig. 2A,B). In this case both CtBP and NCoR were then rapidly dismissed with the progress of the following cycle of ER activation and recruitment (Fig. 2B). While CtBP has not been previously linked directly to estrogen receptor regulation, CtBP is known to be a key component of the repressive functions mediated by the corepressor factor Rip140 (Cavailles et al., 1995; White et al., 2005), which is recruited to ER target genes, such as pS2, in a ligand-dependent fashion (Fig. 2B). Thus, we performed a single cell microinjection experiment to test whether CtBP recruitment by Rip140 is the determining factor imposing the requirement for TBL1 on transcriptional activation. As shown in Figure 2C, removal of endogenous Rip140 by single cell nuclear microinjection of specific αRip140 IgG causes loss of repression. Rescue by overexpressing wild-type Rip140 restores repression and permits TBL1-dependent activation, although the activation is less robust because of the overexpression of Rip140, a potent corepressor, above its endogenous level (Augereau et al., 2006; White et al., 2005). In contrast, if endogenous Rip140 is replaced by overexpressing a mutated Rip140 that is unable to bind CtBP (with a mutation of the main PLDLS motif as described in (Vo et al., 2001)), repression could not be rescued and activation became TBL1-independent (Fig. 2C). These data suggest that Rip140 is required to impose effective CtBP-dependent repression on estrogen receptor target genes, with the requirement for dismissal of the CtBP corepressor complex providing the explanation for the requirement of TBL1 in transcriptional activation.
Figure 2. CtBP1/2 and the NCoR complex are both recruited to ER- and Notch-target genes for repression.
(A) ChIP analysis of the occupancy of the pS2 promoter upon basal growth conditions in U2OS cells showed specific recruitment of CtBP, NCoR, TBLR1 and TBL1 as indicated by qPCR performed on the chromatin immunoprecipitated with each specific antibody compared to the chromatin immunoprecipitated with non-specific IgGs. qPCR of an unrelated genomic region was used as negative control. Results illustrated are representative of three or more biological experiments. (B) Similar ChIP analysis was performed on the pS2 promoter upon estrogen stimulation in MCF7 cells. A peak of recruitment of both CtBP and NCoR could be observed 30’ after ligand stimulation, together with ligand-dependent recruitment of ERα and Rip140, followed by their rapid dismissal at 45’ (C) Single-cell nuclear microinjection of purified IgGs against TBL1 inhibited transcriptional activation of an ERE-dependent LacZ reporter upon E2 in MCF7 cells carrying the endogenous Rip140 or in cells where overexpression of wild-type Rip140 substituted for endogenous Rip140. When a mutant Rip140, unable to bind CtBP, was used to rescue for the loss of endogenous Rip140 the basal repression in absence of stimulation could not be restored and activation became TBL1-independent. (D) ChIP analysis in undifferentiated C2C12 cells (GM) showed enrichment of the Hes1 promoter in the sonicated chromatin fraction immunoprecipitated by CtBP, NCoR and HDAC3 specific antibodies. On the contrary, occupancy of the Hes1 promoter by the Trimethyl-Lys4-histone 3 mark is decreased upon induction of differentiation (DM). Corresponding expression of the Hes1 gene during differentiation is shown in Supplemental Figure S2. (E) The relative induction of Hes1 expression in C2C12 myoblasts upon NICD transient transfection was measured by RT-PCR and normalized for 18S expression. Co-transfection of specific siRNAs against TBL1 and TBLR1 showed that both are required for Notch-mediated transcriptional activation. (F) Single cell microinection assay in C2C12 cells showing that transcriptional activation of a LacZ reporter driven by the Notch-responsive Hes1 promoter requires NICD, CBF, SRC1, CBP, TBL1 and TBLR1.
The CSL (CBF/RBPj-Su(H)-Lag1) family of transcription factors are the primary effectors of Notch signaling in vertebrate and invertebrate organisms. Whereas they function as transcriptional repressors in basal conditions, Notch signaling and nuclear translocation of the NICD intracellular domain converts them in transcriptional activators because of dismissal of corepressor factors and recruitment of coactivators (Mumm and Kopan, 2000). Interestingly, repression of Notch target genes by CSL factors has been reported to be mediated by several specific corepressors, such as CIR, SHARP, Hairless and SKIP, that recruit other general corepressors, including NCoR/SMRT, HDACs, and CtBP (Barolo et al., 2002; Kao et al., 1998; Morel et al., 2001; Mumm and Kopan, 2000). Therefore, we tested the hypothesis that both TBL1 and TBLR1 would be required for Notch signaling activation by investigating the transcriptional activation of one of the most established Notch target gene, the Hairy/Enhancer of split 1 (HES1) gene. To determine whether both corepressor complexes were recruited to the Hes1 promoter region when repressed, we analyzed Hes1 gene expression by RT-PCR in C2C12 cells and observed that its mRNA level was rapidly downregulated upon differentiation from myoblasts to myotubes (Supplemental Fig S2). Accordingly, detection of the trimethyl-K4 histone 3 activation mark on the Hes1 promoter was significantly decreased when ChIP analysis was performed on cross-linked chromatin from cells after 2 days of differentiation compared to undifferentiated C2C12 cells (Fig. 2D). In this same differentiated state, the presence of both CtBP and NCoR/HDAC3 could be recorded on the repressed Hes1 promoter, as expected, but not on an unrelated genomic region (Fig. 2D). Next, we tested whether Notch-dependent activation depends on TBL1 and TBLR1 activity. Because the Hes1 gene is already partly expressed in undifferentiated C2C12 myoblasts, we induced a transient overexpression of the Notch intracellular domain (NICD) to promote a further increase in activation, finding that this increased expression of the endogenous Hes1 gene, as measured by RT-PCR, was dependent on both TBL1 and TBLR1 (Fig. 2E), while a lack of effect of TBL1 and TBLR1 downregulation on basal levels of Hes1 expression probably reflects the low efficiency of transfection in C2C12 cells. Similarly, single cell nuclear microinjection assays, analyzing the expression of a LacZ reporter driven by the Hes1 promoter revealed that the reporter was activated in undifferentiated C2C12 cells, dependent on the presence of the DNA-binding transcription factor, CBF1/CSL, and on a series of coactivators, including SRC-1 and CBP. We found that depletion of either TBL1 or TBLR1 by specific siRNAs completely abrogated transcriptional activation, confirming that both factors are required for Notch-dependent gene activation (Fig. 2F). In conclusion, our data support the hypothesis that negative regulation of target genes of several transcription factors is maintained by the independent efforts of distinct corepressor complexes, in this case NCoR/SMRT and CtBP, and that when such dual repression strategy is enforced the functions of specific exchange factors, here TBL1 and TBLR1, are required.
TBL1 interacts in vivo with CtBP and promote its proteasomal degradation
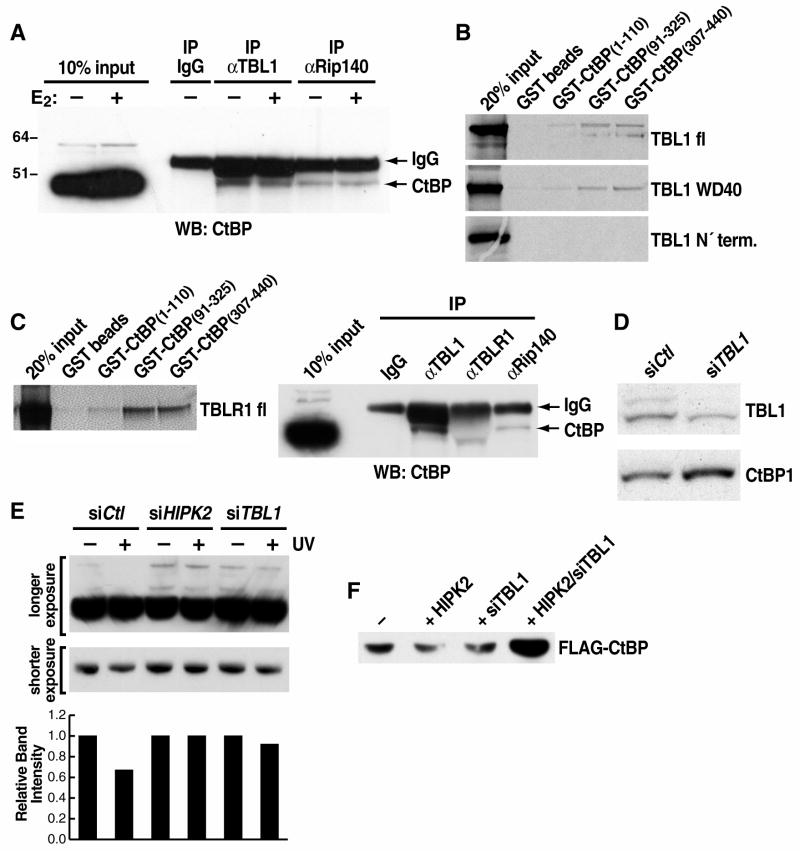
Whereas both TBL1 and TBLR1 are intrinsic components of the NCoR/SMRT complex, no interaction has been previously reported between either of these two cofactors and the CtBP complex. However, based on our previous observations regarding TBLR1, we considered that TBL1 might similarly act as a specific adaptor for CtBP ubiquitylation and degradation. Accordingly, we observed quite a robust in vivo interaction between endogenous TBL1 and CtBP proteins by co-immunoprecipitation in 293 cells, with the interaction of Rip140 with CtBP providing a positive control (Fig. 3A). GST pull-down assay revealed direct interactions between TBL1 and CtBP, which were mapped to the WD40 domain of TBL1 and the C’-terminus of CtBP (Fig. 3B). Interestingly, while an in vitro interaction between CtBP and TBLR1 could be detected, these two proteins interacted very poorly by coimmunoprecipitation (Fig. 3C), suggesting that, in vivo, the strongest interaction occurs between CtBP and TBL1.
Figure 3. TBL1 function in mediating CtBP degradation.
(A) Endogenous co-immunoprecipitation of CtBP with TBL1 and Rip140 performed in estradiol-stimulated MCF-7 cells. Co-immunoprecipitation of CtBP and Rip140 was used as positive control. (B) GST pull down analysis revealing direct interaction between CtBP C’terminus and TBL1 WD40 domain. (C) Direct interaction between TBLR1 and CtBP in a GST pull down assay. In vivo interaction between CtBP and TBL1 was also observed in 293 cells, while co-immunoprecipitation between CtBP and TBLR1 could not be recorded. Immunoprecipitation of NCoR (see Supplemental Figure S3-A) was performed in parallel as positive control. (D) Immunoblot of U2OS whole cell extracts with α-CtBP antibody revealed that TBL1 downregulation by siRNA transfection stabilized CtBP protein level. (E) Immunoblot analysis of extracts from U2OS treated with 50J/cm3 UV showed that CtBP protein degradation is impaired in cells transfected with specific siRNAs against TBL1 or HIPK2. Changes in CtBP protein level is better appreciated on the lower exposure or on the slower-running bands representing post-transcriptionally modified CtBP, as confimed by specific siRNA downregulation (see Supplemental Figure S3-B). Quantification of the CtBP bands from the shorter exposed panel was done using the GeneTools software on a GeneGenius Byo Imaging System and is represented in the bar graph below. (F) Immunoblot with αFLAG of 293 cell extracts upon transient transfection with Flag-CtBP and HA-HIPK2 confirming that CtBP protein level is downregulated by HIPK2 overexpression in a TBL1-dependent manner.
Consistent with the hypothesis that TBL1 functions during transcriptional activation to promote dismissal and degradation of CtBP by recruitment of ubiquitylation machinery, CtBP protein levels were also stabilized in both U2OS and 293 cells when TBL1 was downregulated by specific siRNA (Fig. 3D and data not shown). Similarly, CtBP protein levels were higher in embryonic stem cells null for TBL1 (TBL1Δ/Y) compared to wild-type cells (data not shown). CtBP is known to be ubiquitylated and degraded in response to UV-induced activation of the p53 signaling pathway in a HIPK2-dependent fashion (Zhang et al., 2003). Accordingly, CtBP stabilization in the absence of TBL1 was observed not only on the basal protein level, but also upon UV stress (Fig. 3E) or when protein degradation was specifically induced by HIPK2 overexpression (Fig. 3F) (Zhang et al., 2005; Zhang et al., 2003). As a positive control, CtBP stabilization was also observed with downregulation of HIPK2 by specific siRNA (Fig. 3F). In conclusion, these data together suggest that TBL1 is a CtBP-interacting partner that functions as a specific adaptor for its ubiquitylation and degradation in response to different stimuli.
TBL1 and TBLR1 are specifically regulated by phosphorylation
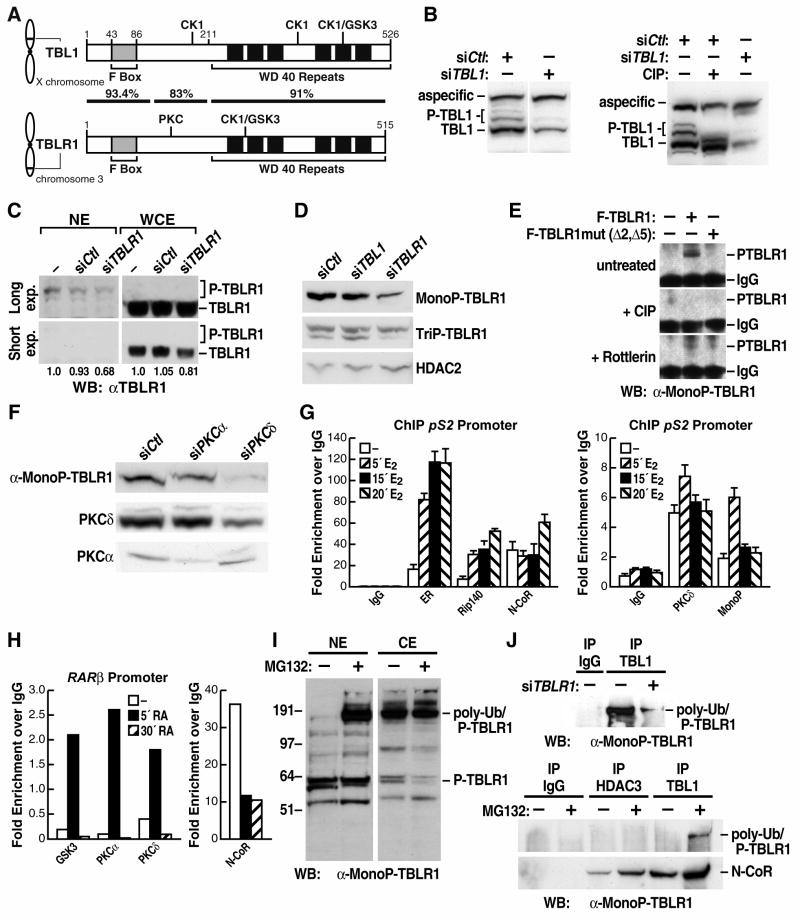
Despite the striking fundamental mechanistic differences between TBL1 and TBLR1 functions, these two factors exhibit very limited differences at the primary amino acid sequence level. Intriguingly, alignment of the protein sequences revealed that almost all of the non-conserved residues fall over five putative phosphorylation sites, each of which is selectively present in one of the two proteins (Supplement Fig. S6). Three putative phosphorylation sites for CK1 and GSK3 were identified by NetPhos prediction analysis as specifically present on TBL1 but not on TBLR1 (S173/T176, T333, S420/S424). Conversely, TBLR1 contained one site for CK1/GSK3 (S199/T203/S204) and one site for PKCδ (S123), both specifically absent in the mammalian TBL1 sequences (Schematic in Fig. 4A). Because of this potentially functional sequence distinction, we analyzed TBL1 and TBLR1 post-transcriptional modifications in vivo. Western blotting with the αTBL1- specific IgGs previously raised in guinea pigs (Perissi et al., 2004) indicated that there were multiple specific bands recognized by the antibody, running at a slightly higher molecular weight than the main TBL1 polypeptide (Fig. 4B). These bands were specifically removed by TBL1 siRNA and shifted by treating the protein extracts with calf intestinal alkaline phosphatase (Fig. 4B). Similar results could be observed for TBLR1 when using nuclear extract (Fig. 4C and data not shown), suggesting that both proteins were phosphorylated at multiple sites in vivo and that the phosphorylated proteins were enriched in the nuclear fraction.
Figure 4. TBL1X and TBLR1 are specifically regulated by phosphorylation.
(A) Schematic representation of murine TBL1 and TBLR1 showing the chromosomal locations of the genes, the position of F-box and WD-40 domains in the translated proteins, the homology between TBL1 and TBLR1 and the localization of five putative phosphorylation sites that are uniquely present on each protein. (B) Immunoblot of whole cell extracts from U2OS cells showing multiple specific bands identified by αTBL1 antibody and removed by siRNA transient transfection. The higher molecular weight bands indicate phosphorylated forms of TBL1 as shown by their depletion with CIP alkaline phosphatase. (C) Immunoblot with αTBLR1 antibody comparing nuclear extracts (NE) and whole cell extracts (WCE) from U2OS cells identified higher molecular weight bands enriched in the nuclear fraction. Quantification of the relative intensity of the bands from the longer exposed blot for the NE and the shorter exposed blot for the WCE is shown below. (D) Immunoblot with α-MonoP-TBLR1 and α-TriP-TBLR1 antibodies on U2OS nuclear extracts confirmed that TBLR1 is phosphorylated in vivo in the nuclear compartment. The antibodies specificity is confirmed by TBLR1 and TBL1 siRNAs; αHDAC2 blotting is used as loading control. (E) Depletion of the immunoprecipitated phospho-TBLR1 from nuclear extracts with CIP phosphatase and treatment of cells with Rottlerin confirmed in vivo phosphorylation by PKC. (F) Immunoblot of nuclear extracts from U2OS transiently transfected with siRNAs against PKCα or PKCδ confirmed that PKCδ phosphorylates TBLR1 at Ser 123, as measured by western blot with the α-MonoP-TBLR1. Specificity of the siRNAs is confirmed by immunoblotting for PKCα and PKCδ (G) ChIP analysis of the occupancy of the pS2 promoter by ERα, Rip140, NCoR and Ser123-phospho-TBLR1 in MCF7 cells treated with estrogen for the indicated times. (H) Occupancy of the RARβ promoter in 293 cells showing transient recruitment of PKCα, PKCδ and GSK3 kinases and dismissal of NCoR 5’ after treatment with retinoic acid. (I) Immunoblot with α-MonoP-TBLR1 antibody showing enrichment of nuclear poly-ubiquitylated phospho-TBLR1 when proteasome activity is blocked by MG132. (J) Poly-ubiquitylated phospho-TBLR1 is immunoprecipitated by endogenous TBL1 and specifically depleted by TBLR1 siRNA transfection in 293 cells. The immunoprecipitation with TBL1 is increased by MG132 while no interaction between poly-ubiquitylated phosphoTBLR1 and HDAC3 is detected. Immunoprecipitation of NCoR is shown as positive control.
We therefore raised phosphopeptide-specific, site-specific antibodies against the two phosphorylated residues that characterized the distinction between TBLR1 versus TBL1. Specifically, a Mono-P-TBLR1 antibody was raised against a 20-aa peptide including phospho-S123, and a TriP-TBLR1 antibody was raised against a 20-aa peptide phosphorylated at S199/T203/S204 (See schematic in Fig. 4A). Both of these IgGs could identify in the nuclear lysates specific bands partially overlapping and corresponding to different combinations of post-translational modifications (Fig. 4D and data not shown). In addition, the major bands identified in western blotting by the phospho-specific TBLR1 antibodies were specifically diminished when U2OS cells were transfected with TBLR1, but not with TBL1 siRNA, confirming factor specificity (Fig. 4D). Immunostaining of U2OS cells with the phospho-specific antibodies also confirmed the nuclear localization of the phosphorylated protein (data not shown). Together, these data confirmed the existence in vivo of nuclear TBLR1 phosphorylated at multiple residues. We next focused our analysis on the characterization of the Mono-P-TBLR1 antibody raised against the putative PKC phosphorylation site. This antibody specifically recognized the wild-type TBLR1 when immunoprecipitated using α-Flag or α-Myc antibody, while it did not recognize TBLR1 when mutated at the specific site of putative PKC phosphorylation (“Δ5”) either alone or in combination with the putative CK1/GSK3 site (“Δ2)” (Supplemental Fig. S4). Consistent with the antibody specificity for the phosphorylated residue, the epitope specifically recognized by the Mono-P-TBLR1 antibody could be eliminated when cell lysates were treated with alkaline phosphatase, and could be greatly diminished by treating cells with a PKCδ-specific inhibitor, Rottlerin, prior to protein extraction (Fig. 4E). Accordingly, in nuclear extract from 293 cells transiently transfected with PKCδ -specific siRNA the amount of phospho-TBLR1 identified by the Mono-P-TBLR1 antibody was greatly diminished (Fig. 4F). Because no change was observed when PKCα was downregulated by specific siRNA, we conclude that TBLR1 is directly phosphorylated in vivo by PKCδ at Ser123 (Fig. 4F).
We next performed ChIP analysis on the promoter of the ERα-regulated pS2 gene, in MCF7 cells, and we discovered that the presence of TBLR1 phosphorylated at Ser123 was significantly enriched on this specific location within 5 minutes following hormonal stimulation (Fig. 4G), suggesting that phosphorylation actually occurs locally on the promoter. To investigate this hypothesis, we tested occupancy of the pS2 promoter by PKCδ and found that it also exhibited a rapid, ligand-dependent recruitment (Fig. 4G). Similarly, we observed rapid ligand-induced recruitment of both PKCδ and GSK3 to the RARβ promoter in 293 cells (Fig. 4H), as previously reported (Kambhampati et al., 2003), in parallel to NCoR dismissal (Fig. 4H). In conclusion, our results suggest that TBLR1 is specifically phosphorylated by PKCδ in situ, on regulated promoters, following stimulation. In additon, we speculate that the modification of TBLR1 is quite transient, as phospho-TBLR1 is rapidly poly-ubiquitylated and degraded, possibly to allow for recycling and recruitment of a new NCoR/HDAC3/TBL1/TBLR1 corepressor complex. The robustness of the Mono-P-TBLR1 antibody was in fact greatly enhanced in the presence of the MG132 proteosome inhibitor, permitting the identification of poly-ubiquitylated, phosphorylated TBLR1 that would accumulate in the nuclear fraction (Fig. 4I). This polyubiquitylated phospho-TBLR1 was specifically eliminated by TBLR1 siRNA and could be immunoprecipitated with endogenous TBL1, but not with the NCoR/HDAC3 complex (Fig. 4J), suggesting that these modifications occurred during the activation process.
Molecular determinants of TBL1 and TBLR1 specificity
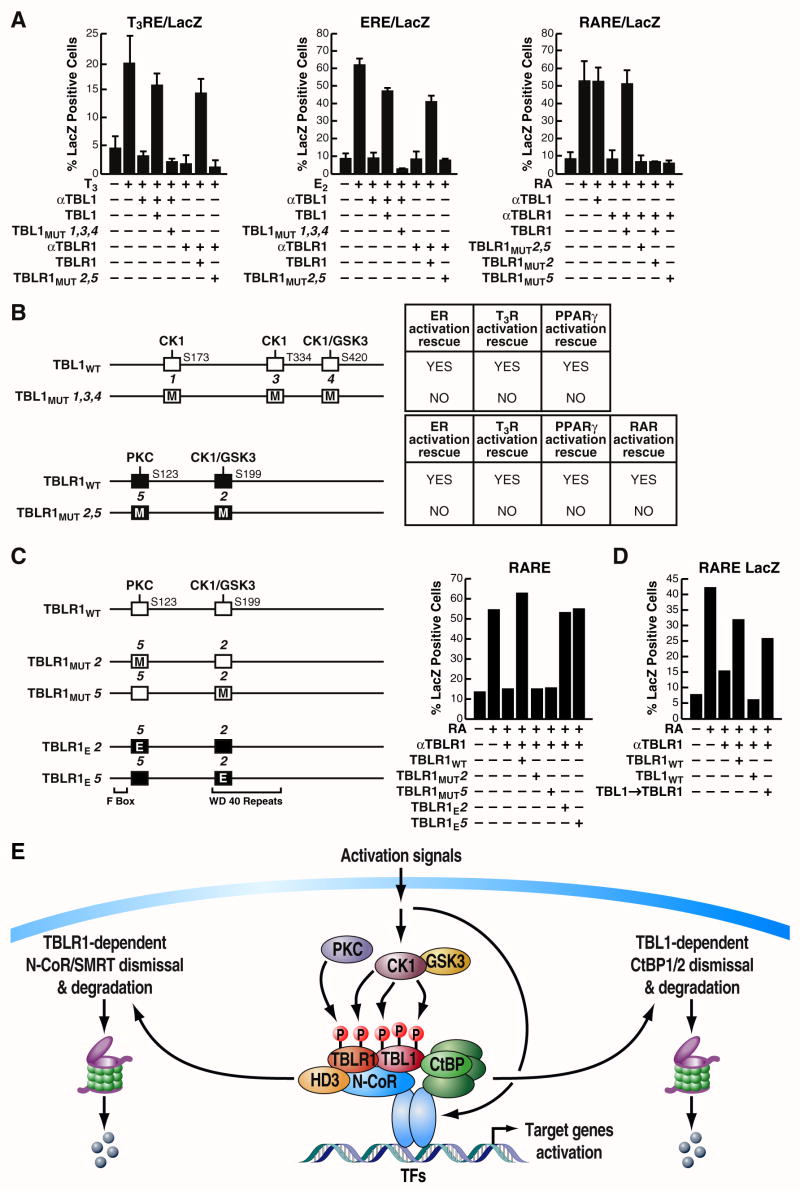
Because of this intriguing distinction between TBL1 and TBLR1 post-translational modifications, we next investigated the relevance of the putative phosphorylation sites for the actions of TBL1 and TBLR1, and for their ability to mediate specific functions. First, we introduced specific mutations in TBL1 and TBLR1 expression vectors, converting each of these sites to the corresponding residues on the other factor (Schematic in Fig. 5B). By testing the ability of these expression vectors to rescue transcriptional activation that had been inhibited by microinjection of 3 α-TBL1 or α-TBLR1 IgGs (Fig. 5A), we found that TBL1 required its three putative phosphorylation sites to mediate estrogen, thyroid hormone or PPARγ receptor function, whereas the putative phosphorylation sites of TBLR1 were required for its action on T3R, ER, PPARγ, as well as RAR (Fig. 5A and data not shown). As summarized in Figure 5B, our data revealed that differential phosphorylation sites represent key molecular determinants of TBL1 and TBLR1 identity and play a fundamental role in the regulation of their function in gene activation. To examine in further detail the role of these putative phosphorylation sites on gene activation we focused on RAR-mediated gene transcription, which is dependent on TBLR1, but not TBL1 (Perissi et al., 2004), and performed another complementation experiment in which the removal of TBLR1 by microinjection was rescued with a series of equally expressing TBLR1 expression vectors mutagenized at the two phosphorylation sites that characterize TBLR1: S123 and S199/T203/S204. Again, if the residues are mutated to the corresponding residues on TBL1, abolishing in this way the putative phosphorylation sites, the mutated protein product was unable to rescue transcriptional activation (Fig. 5C). However, when the same residues are mutated to glutamic acid (E) to mimick phosphorylation, the mutant TBLR1 was able to rescue transcriptional activation equally well as the wild type protein, confirming that phosphorylation of these specific residues is important for the function of TBLR1. Next, we investigated, in the same experimental setting, whether these residues are not only required for TBLR1 functions but also important to determine the specificity between TBLR1 and TBL1. As expected, transcriptional activation by liganded RAR was impaired when the endogenous TBLR1 was downregulated by siRNA microinjection and could be restored only by overexpression of TBLR1, but not by wild-type TBL1 (Fig. 5D). However, overexpression of a TBL1 expression vector that had been mutated by swapping all five putative phosphorylation sites, so that it now led to the same phosphorylation pattern of TBLR1, was also able to rescue the activation (Fig. 5D). In conclusion, these results suggest that the five phosphorylation sites identified are, surprisingly, the sole determinants of the specificity between TBL1 and TBLR1 on the specific transcription units tested.
Figure 5. TBL1 and TBLR1 identities and functions are determined by five specific phosphorylation sites.
(A) Inhibition of T3R, ER and RAR dependent transcriptional activity by αTBL1 or αTBLR1 was rescued by co-injecting expression vectors for either TBL1 or TBLR1, while activation could not be rescued when the specific phosphorylation sites, selectively present in the two factors, were mutated. (B) Schematic representation of the results of the experiments shown in (A) and others not shown: activation by ER, T3R and PPARγ is blocked by αTBL1 microinjection and rescued by overexpression of TBL1 wild-type but not by the mutated form. Similar results for TBLR1 rescue on ER-, T3R-, PPARγ- and RAR-mediated transcription. The results obtained are marked as positive when at least 60% of the original activity is rescued. (C) Activation of a LacZ reporter driven by the RARE response element is inhibited by TBLR1 siRNA microinjection and can be rescued by wild-type TBLR1 or phosphorylation mimicking mutants (Glu), but not by mutations deleting each of the two TBLR1-specific phosphorylation sites. (D) Selective requirement of TBLR1 for RAR-mediated transcription can be specifically changed by swapping the five specific phosphorylation sites that distinguish TBL1 and TBLR1. (E) Model. While TBLR1 specifically functions to mediate the dismissal of the NCoR/SMRT/HDAC3 corepressor complex, TBL1 function is key to promoting the ubiquitylation and degradation of the corepressor CtBP based on direct interaction. The specificity between TBL1 and TBLR1 functions is regulated upon activation via local phosphorylation, with TBLR1 being specifically phosphorylated by PKCδ at Ser123.
Discussion
While the opposing effects of coactivators and corepressors on regulated gene transcription are well recognized, understanding the regulatory mechanisms that defend precise, signal dependent gene activation programs based on the rapid ability to reimpose a repression “check point” has been less well understood. The deeper understanding of the critical importance of corepressor complexes for establishing tightly controlled, signal-dependent activation of gene targets is only recently emerging. Here, we have provided evidence that precise, dedicated strategies for preventing errant basal activation, is achieved, in part, by the combinatorial use of distinct corepressor complexes and the reciprocal utilization of complementary “exchange factors”. Intriguingly, the same strategy, based on the combinatorial actions of TBL1 and TBLR1, is adopted for most nuclear receptors, for the NFκB- regulated genes and for targets of the Notch signaling pathway. We describe here an unexpected, direct interaction between TBL1 and the corepressors CtBP1/2, based on which TBL1 can act as a specific adaptor to promote CtBP ubiquitylation and degradation in response to different signals. Intriguingly, CtBP has never been reported in any of the several NCoR/SMRT complexes that have been isolated from different cellular systems, suggesting that components of one repressor complex maybe important for regulating its own dismissal as well as the dismissal of additional repressors, independently recruited. In addition, while the CtBP complex has not previously been clearly related to the actions of estrogen receptor, we now report, based on the physiological actions of the TBL1 exchange factor, its actions as an additional repression checkpoint for ER-mediated transcriptional regulation. Intriguingly, in this system, Rip140 seems to be required for imposing CtBP-dependent repression, consistent with the well-know interactions between CtBP and Rip140 (Augereau et al., 2006; Vo et al., 2001). However, the temporal kinetics of CtBP recruitment to the pS2 promoter does not fully resemble Rip140’s, suggesting that post-translational modifications such as acetylation/deacetylation (Vo et al., 2001) may impact on the local interaction between these two factors or that Rip140 may play additional roles with alternative partners. Furthermore, it is possible that other factors, such as the ligand dependent corepressor LCoR or the ER corepressor Znf366 (Fernandes et al., 2003; Lopez-Garcia et al., 2006), may also participate to impose CtBP repression. Indeed, recent reports have suggested that multiple repressor complexes may be used combinatorially and be recruited by nuclear receptors in a sequential fashion, similarly to what described for numerous coactivator complexes (An et al., 2004; Liu and Bagchi, 2004; Metivier et al., 2003; Perissi and Rosenfeld, 2005; Yoon and Wong, 2006).
While our data support a profound similarity between the actions of TBL1 and TBLR1, it also underlines their specificity regarding the targets of their actions and the transcription factors requiring them. In answer to the fascinating question of how the specific functions of exchange factors are so precisely maintained, we propose this is achieved, at least in part, by the strategy of regulating phosphorylation of exchange factors such as TBL1 and TBLR1 directly at the level of target gene promoters. In particular, we have identified TBLR1 as a specific target of PKCδ phosphorylation, with Ser123 being phosphorylated in situ on ER and RAR target promoters within minutes of induction. Surprisingly, the specificity of the actions of TBL1 for CtBP dismissal and TBLR1 for NCoR/SMRT dismissal seems to reside in only the few specific phosphorylation sites that are distinct between these two exchange factors, as swapping of these sites is sufficient to change their specificity for RAR-mediated transcriptional activation.
The resulting dismissal of restricting corepressor complexes by this local phosphorylation- dependent strategy extends our constantly growing knowledge of the phosphorylation-dependent regulation of gene transcription by diverse signaling pathways (Baek et al., 2002; Barnes et al., 2003; Fernandez-Majada et al., 2007; Hermanson et al., 2002; Hoberg et al., 2004; Wu et al., 2007). Phosphorylation of Ser123 of TBLR1 on regulated gene promoters by PKCδ corresponds to the activation of PKCδ in response to various hormonal stimuli and to its positive effect on estrogen receptor-, retinoic acid receptor-, peroxisome proliferators-activated receptor- and NFκB-mediated gene activation (Blanquart et al., 2004; Boyan et al., 2003; Catley et al., 2004; De Servi et al., 2005; Ochoa et al., 2003; Radominska-Pandya et al., 2000). Furthermore, the existence of a local kinase regulatory network is consistent with a large number of kinases that have been recently recorded on target gene promoters or even associated with the chromatin of entire transcription units (Chow and Davis, 2006).
In conclusion, we have reported that a critical component of regulated gene transcription is the coordinate actions of TBL1 and TBLR1 in response to distinct ligands and signaling pathways to dismiss distinct corepressor complexes and we believe that this mechanism is important to ensure the physiological amplitude of signal-dependent gene transcription. In addition to the well-described direct phosphorylation of the targets of Fbox-WD40 proteins, we now propose that phosphorylation of the exchange factors themselves is used as a key local switch to permit regulated dismissal of distinct repression “checkpoints” via diverse corepressor complexes.
Experimental Procedures
Reagents, Antibodies and Cell Culture
Anti-TBL1 and anti- TBLR1 antibodies have been described previously (Perissi et al., 2004). Anti-monophospho- and anti-triphospho-TBLR1 antibodies were generated in rabbit against the specific peptides CNQQGS(P)AKNG and CWNLS(P)ENST(P)S(P)GPTQ respectively (PhosphoSolutions). The following antibodies were obtained from Santa Cruz Biotechnology: anti-CtBP (C1 and H440), anti-c-Myc (9E10), anti-ER (MC-20 and H184), anti-Rip140 (H300), anti-HDAC2 (C8) and anti-HDAC3 (H99). Rabbit anti-N-CoR (Jepsen et al., 2000), mouse anti-Flag M2 and mouse anti-β-tubulin (Sigma) were also used in immunoprecipitation and immunoblotting experiments. The PKCδ inhibitor Rottlerin (Santa Cruz) was used at 1uM final concentration. Standard molecular cloning and tissue culture were performed as described by J. Sambrook, D. W. Russell, Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., ed. 3rd, 2001). C2C12 murine myoblast cells were grown in DMEM with 10%FCS until they reached 80% confluency and then switched to DMEM supplemented with 2%HS for the time indicated.
Protein extraction and Immunoprecipitation
For nuclear protein extraction, the cells were rinsed in PBS, harvested and lysed in hypotonic buffer (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 1 mM DTT, 2mM Na2VO3, 50mM NaF, 0.5mM PMSF and protease inhibitor mix, Roche) for 10 minutes on ice, followed by incubation of the nuclear pellet for 20 min in high-salt buffer (10 mM Hepes pH 7.9, 20% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2mM EDTA, 0.5mM DTT, 2mM Na3VO4, 50mM NaF, 1mM PMSF and protease inhibitor mix). For immunoprecipitation and whole cell extracts preparation, cells were rinsed in PBS, harvested and incubated for 20’ on ice in IPH buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 2 mM Na2VO3, 1mM PMSF and protease inhibitor mix. In both cases, concentration of protein extracts was measured using the colorimetric BIORAD assay and the extracts were either boiled in SDS sample buffer and loaded 10% Bistris NuPAGE gel (Invitrogen) or used for immunoprecipitation.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described (Perissi et al., 2004). MCF7 cells were grown in red-free DMEM with 10% charcoal-stripped medium for 72 hours and then treated with 100 nM estradiol; U2OS and 293 cells were serum starved for 24–48h and treated with 10−6M retinoic acid for indicated times. Immunoprecipitated DNA was purified with QIAquick spin colums (Qiagen) and analyzed by qPCR (Stratagene) using Sybergreen, with each PCR performed in triplicate. The percentage of immunoprecipitated chromatin was calculated from the ΔCt values for each sample relative to a curve performed for each experiment with the input chromatin prior to immunoprecipitation. Amplification of unrelated genomic regions was used as negative control and the percentage of specific immunoprecipitated chromatin was converted to fold changes over respective mock immunoprecipitation. The primers used to amplify the pS2 and the RAR-beta promoters have been described previously (Perissi et al., 2004).
GST-affinity purification and pull-down
Fusion proteins containing GST bound to different regions of CtBP1 (CtBP A: aa 1–110; CtBP B: aa 91–325; CtBP C: aa 307–440) were expressed in BL21 bacteria and purified from homogenized lysates with glutathione-agarose beads (Sigma), as previously described (Perissi et al., 2004). For interaction studies, the immobilized GST-fusion proteins were incubated with 35S-labeled TBL1 full-length or segments (WD4 domain: aa 211–526; N’ terminal: aa 1–211), obtained by in vitro translation-transcription (Promega TnT Quick Coupled Transcription/Translation System), followed by extensive washes, separation of the interacting proteins by SDS-PAGE and autoradiography.
Single cell nuclear microinjection assay and transient transfection
The single cell nuclear microinjection assays were performed as described in McInerney et al., 1998. Each experiment was performed on three independent coverslips with >300 injected cells per point, and rhodamine-conjugated dextran was used as a negative control in each experiment. Before injection, cells were rendered quiescent by incubation in serum-free medium for 24–36h. The LacZ reporters have all been described previously (McInerney et al., 1998; Sheppard et al., 1999: Perissi et al., 2004). Transient transfection experiments were performed with Lipofectamine 2000 (Promega). Murine clones of TBL1 and TBLR1 were subcloned into pCMX-HA-FLAG-tag and into pCS2-Myc-tag expression vectors. The mutants lacking the kinases recognition sites or mimicking constitutive activation were obtained by site-specific mutagenesis of the indicated serine/threonine residues. Specific siRNAs against hTbl1X (1255–1275) and hTblR1 (1634–1654) were designed (Qiagen) and removal of gene expression was confirmed by RT-PCR.
RNA Isolation and RT-PCR Analysis
RNA was isolated following the manufacturer protocol for the RNeasy Kit (QIAGEN). First strand cDNA synthesis from total RNA template was performed with Invitrogen Superscript III cDNA Synthesis System, followed by SYBR-green qPCR amplification. Normalization was performed using specific amplification of GAPDH and 18S RNA and PCRs were performed in triplicate for each biological duplicate experiment.
Supplementary Material
Acknowledgments
We are grateful to C.Nelson and A.Krones for their excellent technical assistance and we deeply appreciate the helpful insights and discussions from all the members of the Rosenfeld’s and Glass’s laboratories. We gratefully acknowledge Dr. A. Aggarwaal for protein modeling and for his helpful advices. We thank J. Hightower for help with the artwork and figure preparation, K. Nixon at PhosphoSolutions for antibodies preparation and Dr. L.Puri for C2C12 cells. V.P. is supported by NIDDK (grant K99DK078756), C.S. is supported by AICF, M.G.R. is an HHMI investigator and these studies were supported by the following NIH grants: NS34934, DK39949, DK74868, DK018477.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Asahara H, Dutta S, Kao HY, Evans RM, Montminy M. Pbx-Hox heterodimers recruit coactivator-corepressor complexes in an isoform-specific manner. Mol Cell Biol. 1999;19:8219–8225. doi: 10.1128/mcb.19.12.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augereau P, Badia E, Balaguer P, Carascossa S, Castet A, Jalaguier S, Cavailles V. Negative regulation of hormone signaling by RIP140. J Steroid Biochem Mol Biol. 2006;102:51–59. doi: 10.1016/j.jsbmb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Barnes CJ, Vadlamudi RK, Mishra SK, Jacobson RH, Li F, Kumar R. Functional inactivation of a transcriptional corepressor by a signaling kinase. Nat Struct Biol. 2003;10:622–628. doi: 10.1038/nsb957. [DOI] [PubMed] [Google Scholar]
- Barolo S, Stone T, Bang AG, Posakony JW. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 2002;16:1964–1976. doi: 10.1101/gad.987402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanquart C, Mansouri R, Paumelle R, Fruchart JC, Staels B, Glineur C. The protein kinase C signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor alpha. Mol Endocrinol. 2004;18:1906–1918. doi: 10.1210/me.2003-0327. [DOI] [PubMed] [Google Scholar]
- Boyan BD, Sylvia VL, Frambach T, Lohmann CH, Dietl J, Dean DD, Schwartz Z. Estrogen-dependent rapid activation of protein kinase C in estrogen receptor-positive MCF-7 breast cancer cells and estrogen receptor-negative HCC38 cells is membrane-mediated and inhibited by tamoxifen. Endocrinology. 2003;144:1812–1824. doi: 10.1210/en.2002-221018. [DOI] [PubMed] [Google Scholar]
- Catley MC, Cambridge LM, Nasuhara Y, Ito K, Chivers JE, Beaton A, Holden NS, Bergmann MW, Barnes PJ, Newton R. Inhibitors of protein kinase C (PKC) prevent activated transcription: role of events downstream of NF-kappaB DNA binding. J Biol Chem. 2004;279:18457–18466. doi: 10.1074/jbc.M400765200. [DOI] [PubMed] [Google Scholar]
- Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner PJ, Parker MG. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. Embo J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol Cell. 2002;9:213–224. doi: 10.1016/s1097-2765(02)00443-4. [DOI] [PubMed] [Google Scholar]
- Chow CW, Davis RJ. Proteins kinases: chromatin-associated enzymes? Cell. 2006;127:887–890. doi: 10.1016/j.cell.2006.11.015. [DOI] [PubMed] [Google Scholar]
- De Servi B, Hermani A, Medunjanin S, Mayer D. Impact of PKCdelta on estrogen receptor localization and activity in breast cancer cells. Oncogene. 2005;24:4946–4955. doi: 10.1038/sj.onc.1208676. [DOI] [PubMed] [Google Scholar]
- Fernandes I, Bastien Y, Wai T, Nygard K, Lin R, Cormier O, Lee HS, Eng F, Bertos NR, Pelletier N, et al. Ligand-dependent nuclear receptor corepressor LCoR functions by histone deacetylase-dependent and -independent mechanisms. Mol Cell. 2003;11:139–150. doi: 10.1016/s1097-2765(03)00014-5. [DOI] [PubMed] [Google Scholar]
- Fernandez-Majada V, Aguilera C, Villanueva A, Vilardell F, Robert-Moreno A, Aytes A, Real FX, Capella G, Mayo MW, Espinosa L, Bigas A. Nuclear IKK activity leads to dysregulated notch-dependent gene expression in colorectal cancer. Proc Natl Acad Sci U S A. 2007;104:276–281. doi: 10.1073/pnas.0606476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Barak O, Lazar MA. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–6101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hermanson O, Jepsen K, Rosenfeld MG. N-CoR controls differentiation of neural stem cells into astrocytes. Nature. 2002;419:934–939. doi: 10.1038/nature01156. [DOI] [PubMed] [Google Scholar]
- Hoberg JE, Yeung F, Mayo MW. SMRT Derepression by the IkappaB Kinase alpha; A Prerequisite to NF-kappaB Transcription and Survival. Mol Cell. 2004;16:245–255. doi: 10.1016/j.molcel.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Kambhampati S, Li Y, Verma A, Sassano A, Majchrzak B, Deb DK, Parmar S, Giafis N, Kalvakolanu DV, Rahman A, et al. Activation of protein kinase C delta by all-trans-retinoic acid. J Biol Chem. 2003;278:32544–32551. doi: 10.1074/jbc.M301523200. [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Kohlbrecher D, Adhikary S, Eilers M. Mechanisms of transcriptional repression by Myc. Curr Top Microbiol Immunol. 2006;302:51–62. doi: 10.1007/3-540-32952-8_3. [DOI] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Lee SK, Kim JH, Lee YC, Cheong J, Lee JW. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappaB, and serum response factor. J Biol Chem. 2000;275:12470–12474. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Liu XF, Bagchi MK. Recruitment of distinct chromatin-modifying complexes by tamoxifen-complexed estrogen receptor at natural target gene promoters in vivo. J Biol Chem. 2004;279:15050–15058. doi: 10.1074/jbc.M311932200. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia J, Periyasamy M, Thomas RS, Christian M, Leao M, Jat P, Kindle KB, Heery DM, Parker MG, Buluwela L, et al. ZNF366 is an estrogen receptor corepressor that acts through CtBP and histone deacetylases. Nucleic Acids Res. 2006;34:6126–6136. doi: 10.1093/nar/gkl875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiakowski P, Breathnach R, Bloch J, Gannon F, Krust A, Chambon P. Cloning of cDNA sequences of hormone-regulated genes from the MCF-7 human breast cancer cell line. Nucleic Acids Res. 1982;10:7895–7903. doi: 10.1093/nar/10.24.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Kopan R. Notch signaling: from the outside in. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- Ochoa WF, Torrecillas A, Fita I, Verdaguer N, Corbalan-Garcia S, Gomez-Fernandez JC. Retinoic acid binds to the C2-domain of protein kinase C(alpha) Biochemistry. 2003;42:8774–8779. doi: 10.1021/bi034713g. [DOI] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A, Chen G, Czernik PJ, Little JM, Samokyszyn VM, Carter CA, Nowak G. Direct interaction of all-trans-retinoic acid with protein kinase C (PKC). Implications for PKC signaling and cancer therapy. J Biol Chem. 2000;275:22324–22330. doi: 10.1074/jbc.M907722199. [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y, Shi Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Tomita A, Buchholz DR, Shi YB. Recruitment of N-CoR/SMRT-TBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol Cell Biol. 2004;24:3337–3346. doi: 10.1128/MCB.24.8.3337-3346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Fjeld C, Goodman RH. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol Cell Biol. 2001;21:6181–6188. doi: 10.1128/MCB.21.18.6181-6188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Yore MM, Deng D, Spinella MJ. Limiting effects of RIP140 in estrogen signaling: potential mediation of anti-estrogenic effects of retinoic acid. J Biol Chem. 2005;280:7829–7835. doi: 10.1074/jbc.M412707200. [DOI] [PubMed] [Google Scholar]
- Wong CW, Privalsky ML. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARalpha, and BCL-6. J Biol Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- Wu RC, Feng Q, Lonard DM, O’Malley BW. SRC-3 Coactivator Functional Lifetime Is Regulated by a Phospho-Dependent Ubiquitin Time Clock. Cell. 2007;129:1125–1140. doi: 10.1016/j.cell.2007.04.039. [DOI] [PubMed] [Google Scholar]
- Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen TM, Heinzel T, Szeto D, Korzus E, Kurokawa R, et al. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Huang ZQ, Li J, Fondell JD, Qin J, Wong J. Purification and functional characterization of the human N-CoR complex: the roles of HDAC3, TBL1 and TBLR1. Embo J. 2003;22:1336–1346. doi: 10.1093/emboj/cdg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Wong J. The corepressors silencing mediator of retinoid and thyroid hormone receptor and nuclear receptor corepressor are involved in agonist- and antagonist-regulated transcription by androgen receptor. Mol Endocrinol. 2006;20:1048–1060. doi: 10.1210/me.2005-0324. [DOI] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Chait BT, Roeder RG. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002a;9:611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Nottke A, Goodman RH. Homeodomain-interacting protein kinase-2 mediates CtBP phosphorylation and degradation in UV-triggered apoptosis. Proc Natl Acad Sci U S A. 2005;102:2802–2807. doi: 10.1073/pnas.0409373102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002b;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yoshimatsu Y, Hildebrand J, Frisch SM, Goodman RH. Homeodomain interacting protein kinase 2 promotes apoptosis by downregulating the transcriptional corepressor CtBP. Cell. 2003;115:177–186. doi: 10.1016/s0092-8674(03)00802-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.