Abstract
The expression of peroxisome proliferator-activated receptor (PPAR)γ in thyroid neoplasias and in normal thyroid (NT) tissues has not been fully investigated. The objectives of the present work were: to study and compare the relative expression of PPARγ in normal, benign and malignant thyroid tissues and to correlate PPARγ immunostaining with clinical/pathological features of patients with thyroid cancer. We analysed the expression of PPARγ in several types of thyroid tissues by reverse transcription–polymerase chain reaction (RT–PCR), interphase fluorescent in situ hybridisation, real-time RT–PCR and immunohistochemistry. We have demonstrated that NT tissues express PPARγ both at mRNA and at protein level. PAX8-PPARγ fusion gene expression was found in 25% (six of 24) of follicular thyroid carcinomas (FTCs) and in 17% (six of 36) of follicular thyroid adenomas, but in none of the 10 normal tissues, 28 nodular hyperplasias, 38 papillary thyroid carcinomas (PTCs) and 11 poorly differentiated thyroid carcinomas (PDTCs). By real-time RT–PCR, we observed that tumours negative for the PAX8-PPARγ rearrangement expressed lower levels of PPARγ mRNA than the NT. Overexpression of PPARγ transcripts was detected in 80% (four of five) of translocation-positive tumours. Diffuse nuclear staining was significantly (P<0.05) less prevalent in FTCs (53%; 18 of 34), PTCs (49%; 19 of 39) and PDTCs (0%; zero of 13) than in normal tissue (77%; 36 of 47). Peroxisome proliferator-activated receptorγ-negative FTCs were more likely to be locally invasive, to persist after surgery, to metastasise and to have poorly differentiated areas. Papillary thyroid carcinomas with a predominantly follicular pattern were more often PPARγ negative than classic PTCs (80% vs 28%; P=0.01). Our results demonstrated that PPARγ is underexpressed in translocation-negative thyroid tumours of follicular origin and that a further reduction of PPARγ expression is associated with dedifferentiation at later stages of tumour development and progression.
Keywords: PPARγ, PAX8, underexpression, follicular thyroid tumours
Peroxisome proliferator-activated receptors (PPARs) are members of the nuclear receptor superfamily, which form heterodimers with retinoid X receptor. The heterodimers activate the transcription of specific genes in response to binding of a ligand. Three PPAR isoforms have been described: α, β (also called δ, NUC-1 or FAAR) and γ (Desvergne and Wahli, 1999). PPARγ is the most intensively studied isoform. It has been shown that this nuclear receptor is important in several biological pathways involving cell differentiation, insulin sensitivity, atherosclerosis and cancer (Rosen and Spiegelman, 2001). There are two protein isoforms (PPARγ1 and PPARγ2) generated by alternative splicing and alternative promoter usage. Peroxisome proliferator-activated receptorγ1 isoform is encoded by three transcripts, which differ in 5′-untranslated region (variants γ1, γ3 and γ4). Variant γ2 encodes isoform PPARγ2 (Martin et al, 1998; Sundvold and Lien, 2001). Peroxisome proliferator-activated receptorγ2 contains 30 additional amino acids in the N-terminus (Tontonoz et al, 1994). Peroxisome proliferator-activated receptorγ is activated by natural ligands (fatty acids and eicosanoids) (Chawla and Lazar, 1994; Tontonoz et al, 1994; Forman et al, 1995; Kliewer et al, 1995) and by synthetic ligands (thiazolidinediones) (Lehmann et al, 1995). Peroxisome proliferator-activated receptorγ activation was reported to inhibit the growth and, in some cases, to induce apoptosis or differentiation of tumour cells from different lineages: liposarcoma (Tontonoz et al, 1997; Demetri et al, 1999), breast cancer (Elstner et al, 1998; Mueller et al, 1998), prostate cancer (Kubota et al, 1998), colorectal cancer (Brockman et al, 1998; Sarraf et al, 1998; Kitamura et al, 1999), bladder cancer (Guan et al, 1999), non-small-cell lung carcinoma (Chang and Szabo, 2000), pancreatic cancer (Motomura et al, 2000), gastric cancer (Sato et al, 2000), renal carcinoma (Inoue et al, 2001), testicular cancer (Hase et al, 2002) and liver cancer (Toyoda et al, 2002).
Kroll et al (2000) reported that t(2;3)(q13;p25), a chromosomal translocation detected in a subset of follicular thyroid carcinomas (FTCs), originates a fusion gene composed by DNA-binding domain of the thyroid transcription factor PAX8 and domains A to F of PPARγ. Recently, our group and others (Marques et al, 2002; Nikiforova et al, 2002; Cheung et al, 2003) have detected the expression of PAX8-PPARγ gene not only in FTCs but also in follicular thyroid adenomas (FTAs). Ohta et al (2001) studied the expression of PPARγ in papillary thyroid carcinoma (PTC) cell lines and in thyroid tumours. They showed that most cell lines and half of PTCs expressed PPARγ, while normal adjacent tissue and two FTAs were negative. This group as well as Martelli et al (2002) also demonstrated that PPARγ agonists induce apoptosis and inhibit the growth of thyroid tumour cells.
Several studies have demonstrated that, compared to their normal counterparts, the expression of PPARγ in tumour cells is either overexpressed, such as in renal cell carcinoma (Inoue et al, 2001) and testicular cancer (Hase et al, 2002), underexpressed, such as in oesophageal carcinomas (Terashita et al, 2002) or is equal to the normal tissue, such as in colonic adenocarcinomas (Sarraf et al, 1998). This last group has also identified somatic mutations of PPARγ in four of 55 sporadic primary colorectal carcinomas (Sarraf et al, 1999). The expression of PPARγ in thyroid neoplasias and in the normal thyroid (NT) tissue has not been fully investigated. We have expanded our previous study (Marques et al, 2002) and analysed the expression of PPARγ in a series of thyroid tumours and correspondent normal tissue by reverse transcription–polymerase chain reaction (RT–PCR), interphase fluorescent in situ hybridisation (FISH), real-time RT–PCR and immunohistochemistry. We observed that PPARγ expression is usually underexpressed in multiple types of thyroid tumours, and that this may be an important event in the development of thyroid neoplasias.
MATERIALS AND METHODS
Materials
The number of cases analysed by each technique for the different histological groups is represented in Table 1 . Paraffin-embedded tissues and frozen tissues were available in 247 samples and in 131 samples, respectively. Haematoxylin- and eosin-stained sections from each sample were evaluated histologically by two pathologists to classify tumours according to the 1988 World Health Organisation histological classification of thyroid tumours. The extent of papillary carcinomas was classified according to the system of DeGroot et al (1990) and the metastasis-age-completeness-of-resection-invasion-size-score (MACIS) (Hay et al, 1993). The system of DeGroot et al (1990) categorises the patients with PTC by clinical class: I, with intrathyroidal disease; II, with cervical adenopathies; III, with extrathyroidal invasion and IV, with distant metastasis. The prognostic score defined as MACIS was calculated according to Hay et al (1993): MACIS=3.1 (if aged ⩽39 years) or 0.08 × age (if aged ⩾40 years), +0.3 × tumour size (in centimetres), +1 (if incompletely resected), +1 (if locally invasive) and +3 (if distant metastasis present).
Table 1. Thyroid tissues analysed for PPARγ expression.
|
Techniques |
|||
|---|---|---|---|
| Histology | RT–PCR/FISH | Real-time RT–PCR | PPARγ staining |
| Normal thyroid | 10 | 10 | 47 |
| Nodular hyperplasia | 28 | 0 | 28 |
| Follicular adenomas | 36 | 13 | 86 |
| Follicular carcinomas | 24 | 7 | 34 |
| Papillary carcinomas | 38 | 9 | 39 |
| Poorly differentiated carcinomas | 11 | 0 | 13 |
RT–PCR=reverse transcription–polymerase chain reaction; FISH=fluorescent in situ hybridisation; PPAR=peroxisome proliferator-activated receptor.
RNA extraction, RT–PCR and sequencing
Total RNA was extracted from frozen tumours using TRIzol reagent (Life Technologies, Inc., Gaithersburg, MD, USA), according to the manufacturer's protocol. RNA was quantified by UV spectrophotometry (optical density measured at 260 nm). Complementary DNA (cDNA) was synthesised from 1 μg of RNA at 37°C for 90 min, using oligo-(dT) primers (Life Technologies, Inc.) and reverse transcriptase (Life Technologies, Inc.). PAX8-PPARγ fusion gene expression was analysed by RT–PCR as described previously (Marques et al, 2002).
To analyse the expression of the various PPARγ mRNA isoforms, segments from the 5′-terminal region of the PPARγ gene were amplified by PCR using forward primers, located in exons A1, A2 and B and reverse primers located in exon 1. Primer sequences are presented in Table 2 . First round amplifications were performed using 1 μl of cDNA, forward primers P1, P3 and P5 and reverse primer P7. A measure of 1 μl of each amplification reaction was then used as template for second amplification reactions with nested primers P2, P4, P6 (forward) and P8 (reverse). A total of 25 μl reactions were carried out on over 35 cycles using the following conditions: 95°C for 1 min, 55–57°C for 1 min and 72°C for 1 min. Amplification reactions contained final concentrations of 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 200 μM dNTPs (Amersham Pharmacia Biotech, Uppsala, Sweden), 1.0–2.5 mM MgCl2, 10 pmol of each primer (forward and reverse) and 1.5 U of Taq DNA Polymerase (Life Technologies, Inc.). Negative controls for cDNA synthesis and PCRs, in which the template was replaced by sterile water, were included in each experiment. RNA integrity and efficiency of cDNA synthesis were tested in each sample by performing RT–PCR amplification for the housekeeping gene phosphoglycerate kinase-1 (Sugg et al, 1998). Normal colon tissue was used as positive control for the analysis of PPARγ expression (Sarraf et al, 1998).
Table 2. RT–PCR oligonucleotide primer sequences.
| Primer | Exon (PPARγ) | Sequence (5′–3′) |
|---|---|---|
| P1 (forward) | A1 | CGGAGCCCGAGCCCGAG |
| P2 (forward) | A1 | CAGCCGCCGCCTGGGGC |
| P3 (forward) | A2 | ACACTAAACCACCAATATACAA |
| P4 (forward) | A2 | CAAGGCCATTTTGTCAAACG |
| P5 (forward) | B | CGGATTGATCTTTTGCTAGAT |
| P6 (forward) | B | GTTATGGGTGAAACTCTGGG |
| P7 (reverse) | 1 | CAAAGGAGTGGGAGTGGTCTa |
| P8 (reverse) | 1 | CATTACGGAGAGATCCACGGTa |
RT–PCR=reverse transcription–polymerase chain reaction; PPAR=peroxisome proliferator-activated receptor.
Kroll et al (2000).
PCR products were analysed and purified by electrophoresis in a 2% agarose gel stained with ethidium bromide. Polymerase chain reaction products were also subjected to automatic sequencing (ABI Prism 310 Genetic Analyser using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit Version 2; Applied Biosystems, PE Corporation, Foster City, CA, USA).
Interphase FISH analysis
Fluorescent in situ hybridisation was performed on isolated nuclei extracted from 50 μm paraffin-embedded sections of thyroid tumours with BAC probes for PPARγ (RPCI 1130 G23, BAC PAC Resources) and PAX8 (RPCI 1165 I12, BAC PAC Resources). Briefly, PPARγ clone DNA was labelled with digoxigenin and PAX8 DNA with biotin by random priming, using the Bioprime DNA labelling system (Invitrogen S.A., Barcelona, Spain). Nuclear suspensions were spotted on SuperFrost slides (Menzel-Glaser, GMbH, Memmert, Germany) and pretreated with 0.1% pepsin (Sigma-Aldrich, St Louis, MO, USA) in 0.2% HCl at 37°C. Probe mixture in 50% formamide in 2 × SSC was codenatured with nuclear DNA at 80°C for 2 min. Detection of the digoxigenin-labelled PPARγ probe was accomplished using an anti-digoxigenin fluorescein antibody (Roche Diagnostics GMbH, Manheim, Germany) and the biotinylated-labelled PAX8 probe with CY3–avidin (Jackson Immunoresearch Lab, West Grove, USA). Nuclei were counterstained with DAPI-Vectashield mounting solution (Vector, Burlingame, USA). Fluorescence hybridisation signals were analysed and recorded with a Cytovision System (Applied Imaging, New Castle, UK). For each case 200 intact nonoverlapping nuclei were counted. Nuclei in which the two probes were fused, touched or were close to each other (distance ⩽1 probe signal) were scored as positive for the fusion gene.
Real-time RT–PCR
The real-time quantitative PCR was performed in a 96-well reaction plate (MicroAmp®Optical 96-Well Reaction Plate, Applied Biosystems, PE Corp.) on an ABI PRISM® 7000 Sequence Detector System (Applied Biosystems, PE Corp.), according to the manufacturer's instructions. TaqMan® One Step PCR Master Mix Reagents Kit (P/N 4309169; Applied Biosystems, PE Corp.) was used to generate fluorescence signals during each PCR cycle. Specific primers and the probe were designed by Pre-Developed Taqman®Assay Reagents (Assays-on-demand, P/N 4331182, Applied Biosystems, PE Corp.). The amplified region contained exons 5 and 6 from PPARγ gene. In order to normalise the differences in the amount of total RNA used in each reaction, we performed the amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA as endogenous control (FG HUMAN GAPDH 0211014, P/N 4333764F, Applied Biosystems, PE Corp.). A pool of five NT tissues was used as calibrator for determining the relative expression of PPARγ gene in the tumour samples as described previously (Lazar et al, 1999).
Immunohistochemistry
Formalin-fixed paraffin-embedded sections (3 μm) were attached to glass slides pretreated with gelatin. The sections were then dried at 37°C overnight and dewaxed with xylol. Endogenous peroxidase was inhibited with 0.6% H2O2 in methanol for 10 min. Antigen retrieval was performed using a stainless-steel 6-l capacity pressure cooker, with 0.01 M sodium citrate buffer (pH 6.0), for 6 min at full pressure. Slides were incubated with normal goat serum 1 : 10 (DAKO X907, DAKO Corp., Golstrup, Denmark) for 10 min before blocking the endogenous avidin and biotin (Vector SP-2001, Vector Laboratories, Inc., Burlingame, CA, USA). Peroxisome proliferator-activated receptorγ primary antibody 1 : 30 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) was incubated for 30 min. Specificity of PPARγ immunostaining was demonstrated by preincubating the samples with PPARγ blocking peptide 1 : 10 (Santa Cruz Biotechnology, Inc.). Bound primary antibody was detected using biotinylated goat anti-mouse and anti-rabbit immunoglobulin G, being subsequently amplified with streptavidin conjugated to horseradish peroxidase (DAKO K5001; DAKO Corp.). All incubations were performed at room temperature. The peroxidase staining reaction was revealed with a solution containing 3,3′-diaminobenzidine tetrachloride. Sections were counterstained with Mayer's haematoxylin, dehydrated and mounted.
Statistical analysis
The frequencies of PPARγ transcript variants and the level of PPARγ mRNA in each tumour histotype were analysed by χ2 test and unpaired t-test, respectively. Peroxisome proliferator-activated receptorγ mRNA levels in PTCs and in corresponding NT tissues were compared using a paired t-test. Peroxisome proliferator-activated receptorγ immunostaining for nodular hyperplasias (NH) and thyroid tumours was compared with the staining in NT tissues by two-tailed Fisher's exact test. We also correlated the PPARγ immunostaining in FTCs and PTCs with clinical/pathological features of the patients by unpaired t-test, two-tailed Fisher's exact test or χ2 test as appropriate. P-values less than 0.05 were considered significant. Statistical analysis was performed using Graph Pad Prism version 2.0 (San Diego, CA, USA).
RESULTS
Analysis of PPARγ transcript variants
RNA from 72 frozen samples (6 normal tissues, 29 FTAs, 9 FTCs, 24 PTCs and four poorly differentiated thyroid carcinomas (PDTCs)) was analysed by RT–PCR. Peroxisome proliferator-activated receptorγ transcript variants were determined by combining the RT–PCR results obtained for each primer pair. Peroxisome proliferator-activated receptorγ3 could be detected only in the five cases that did not present PPARγ1, because RT–PCR with primer pairs P3P7 or P4P8 originated products with exactly the same size in both variants. Most thyroid tissues expressed PPARγ1, PPARγ2 and PPARγ4, and the proportion of specific variants expressed was similar in NT tissues and in the various types of thyroid tumours (data not shown).
PAX8-PPARγ fusion gene expression
The fusion gene was detected by RT–PCR and/or interphase FISH analysis. Six out of 24 (25%) FTCs and six out of 36 (17%) FTAs were positive for PAX8-PPARγ fusion gene expression. The rearrangement was not detected in 10 NT tissues, 28 NHs, 38 PTCs and 11 PDTCs.
Quantitative analysis of PPARγ gene expression
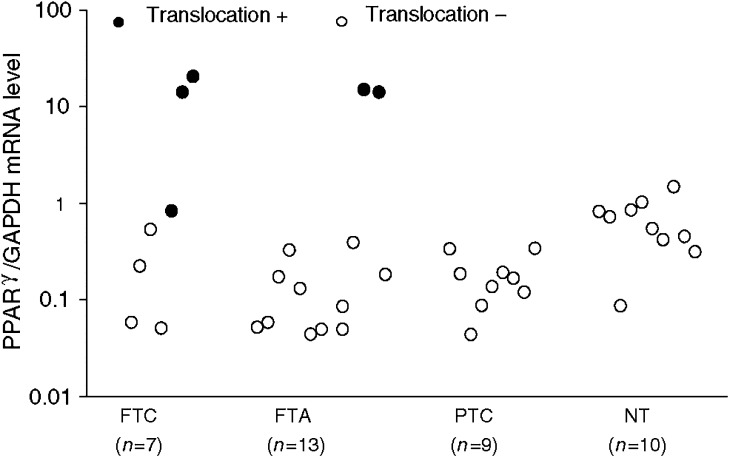
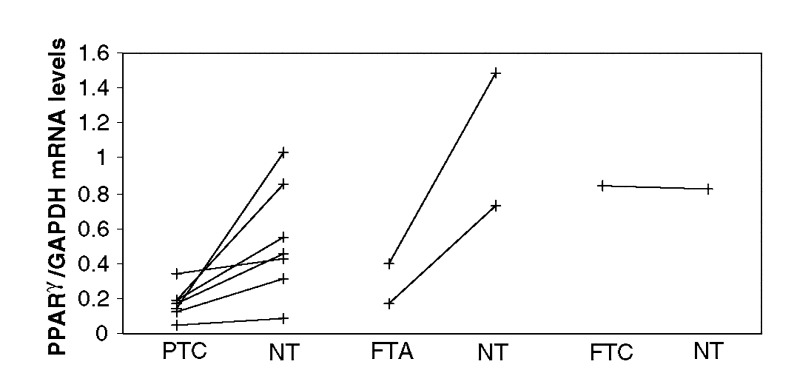
The mRNA level of PPARγ in thyroid tissues is represented in Figure 1. The mean expression level in PAX8-PPARγ-negative FTAs (0.14±0.22; P=0.001) and FTCs (0.22±0.23; P=0.05) was significantly lower than in normal tissue (0.68±0.40). In contrast, PAX8-PPARγ-positive FTAs and FTCs presented mean PPARγ mRNA levels, which were, respectively, 22- (14.68±0.67; P<0.0001) and 17-fold (11.88±10.09; P=0.002), higher than the normal mean. However, one FTC case positive for the fusion gene showed a ratio (0.84) within the normal mean (0.68±0.40). We also observed that PTCs showed a ratio (0.18±0.10; P=0.002) lower than in the NT. This is clearly demonstrated in Figure 2, where paired comparison of PPARγ expression between tumours and normal adjacent tissues from the same patients showed a significant decrease of PPARγ expression in PTCs (0.17±0.09 vs 0.48±0.34; P=0.02).
Figure 1.
Quantitative analysis of PPARγ expression by real-time RT–PCR in several thyroid samples. The mean expression in negative PAX8-PPARγ follicular tumours (FTA: 0.14±0.22; P=0.001 and FTC: 0.22±0.23; P=0.05) and in papillary carcinomas (0.18±0.10; P=0.002) was lower than in NT tissue (0.68±0.40). PAX8-PPARγ-positive follicular adenomas and carcinomas presented PPARγ mRNA levels that were increased by 22- (14.68±0.67; P<0.0001) and 17-fold (11.88±10.09; P=0.002) compared to normal tissue (0.68±0.40). One FTC case positive for the fusion gene exhibited a PPARγ mRNA level (0.84) within the normal mean. FTC – follicular thyroid carcinomas; FTA – follicular thyroid adenomas; PTC – papillary thyroid carcinomas, NT – normal tissue.
Figure 2.
Quantitative analysis of PPARγ mRNA by real-time RT–PCR in thyroid tumours and in the corresponding normal adjacent tissue. The expression level in each PTC was lower than in normal adjacent tissue. The PPARγ mRNA level in two FTAs was also lower than in corresponding normal tissues. One FTC translocation-positive exhibited a ratio similar to its surrounding normal tissue. PTC – papillary thyroid carcinoma; FTA – follicular thyroid adenoma; FTC – follicular thyroid carcinoma; NT – normal thyroid.
PPARγ immunohistochemistry
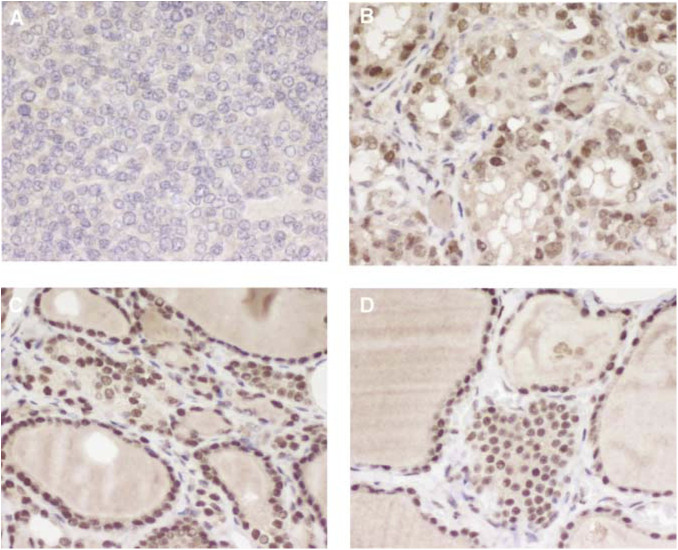
Table 3 presents the intensities of PPARγ nuclear immunostaining for each tumour histotype. Representative results are shown in Figure 3. Diffuse nuclear immunoreactivity, usually faint or moderate, was detected in 77% (36 of 47) of NT tissues, 53% (18 of 34) of FTCs (P=0.03 vs normal tissue), 49% (19 of 39) of PTCs (P=0.01 vs normal tissue), 62% (53 of 86) of FTAs (P=0.09 vs normal tissue) and in 71% (20 of 28) of NHs (P=0.78 vs normal tissue). All PDTCs (n=13) were negative (P<0.0001 vs normal tissue). All translocation-positive FTAs (n=6) and five of six FTCs showed PPARγ staining usually moderate or strong in intensity. Tumour staining was similar to the intensity in the normal adjacent tissue in 62% (29 of 47) of cases, was lower in 28% (13 of 47) of cases and stronger in 10% (five of 47) of cases. Frozen tissue was available in two out of the five cases staining stronger than the normal tissue; PAX8-PPARγ fusion gene was present in these two cases.
Table 3. PPARγ immunohistochemistry in thyroid tumours.
|
PPARγ immunostaining |
|||
|---|---|---|---|
| Tissue | Negative | Positive | P-value |
| NT (n=47) | 11 (23%) | 36 (77%) | |
| NH (n=28) | 8 (29%) | 20 (71%) | NS |
| FTA (n=86) | 33 (38%) | 53 (62%) | NS |
| FTC (n=34) | 16 (47%) | 18 (53%) | 0.03 |
| PTC (n=39) | 20 (51%) | 19 (49%) | 0.01 |
| PDTC (n=13) | 13 (100%) | 0 | <0.0001 |
NS=not significant; NT=normal thyroid; NH=nodular hyperplasia; FTA=follicular thyroid adenoma; FTC=follicular thyroid carcinoma; PTC=papillary thyroid carcinoma; PDTC=poorly differentiated thyroid carcinoma; PPAR=peroxisome proliferator-activated receptor.
Figure 3.
PPARγ protein expression in thyroid tissues assessed by immunohistochemistry. Positive cases presented diffuse nuclear immunostaining. (A) Negative poorly differentiated carcinoma; (B) papillary carcinoma of classic variant with faint immunostaining and corresponding peritumoral (C) and contralateral (D) normal tissue, both with moderate immunoreativity. (A–D) original magnification × 400.
Correlation between PPARγ immunohistochemistry and clinical/pathological data
Clinical and pathological features of malignant tumours are presented in Tables 4 and 5 . When statistical analysis was performed between PPARγ-positive and -negative tumours, we observed that 86% (six of seven) of FTCs with distant metastasis were PPARγ negative (P=0.03). There was also a trend for negative tumours to be more locally invasive (75%), to have poorly differentiated areas (71%) and to have persistent disease after surgery (80%). Most (75%) of the widely invasive FTCs were PPARγ negative, whereas only 38% of minimally invasive tumours did not shown PPARγ staining. The two widely invasive tumours positive for PPARγ had two different components: a follicular area, which stained positive, and an insular area, which was negative. Positive PPARγ staining was not correlated with age, gender, tumour size or vascular invasion. In PTC cases, we observed that 72% (13 of 18) of the tumours with a classic pattern were positive, whereas 80% (12 of 15) of the follicular variants were negative (P=0.01). Peroxisome proliferator-activated receptorγ staining did not correlate with any other prognostic variable but, interestingly, all class IV tumours (n=3) were negative.
Table 4. Clinical and pathological features and PPARγ expression in follicular carcinomas.
|
PPARγ immunohistochemistry |
||||
|---|---|---|---|---|
| FTC | All (n=34) | Negative (n=16) | Positive (n=18) | P-value |
| Age at diagnosis (years±s.d.) | 55.6±16.8 | 59.4±15.7 | 52.2±17.5 | NS |
| F/M ratio | 2.4/1 | 4.3/1 | 1.6/1 | NS |
| Tumour size (cm±s.d.) | 4.1±1.9 | 3.8±1.6 | 4.4±2.2 | NS |
| Invasiveness | NS | |||
| Minimally invasive | 26 | 10 (38%) | 16 (62%) | |
| Widely invasive | 8 | 6 (75%) | 2 (25%) | |
| Poorly differentiated areas | 7 | 5 (71%) | 2 (29%) | NS |
| Local invasion | 4 | 3 (75%) | 1 (25%) | NS |
| Vascular invasion | 24 | 13 (54%) | 11 (46%) | NS |
| Distant metastasis | 7 | 6 (86%) | 1 (14%) | 0.03 |
| Clinical statusa | NS | |||
| Persistent | 5 | 4 (80%) | 1 (20%) | |
| Remission | 28 | 12 (43%) | 16 (57%) | |
| Nonthyroid neoplasias | 3 | 1 (33%) | 2 (67%) | NS |
PPAR=peroxisome proliferator-activated receptor; s.d.=standard deviation; NS=not significant.
Loss of follow-up of one patient.
Table 5. Clinical and pathological features and PPARγ expression in papillary carcinomas.
|
PPARγ immunohistochemistry |
||||
|---|---|---|---|---|
| PTC | All (n=39) | Negative (n=20) | Positive (n=19) | P-value |
| Age at diagnosis (years±s.d.) | 41.0±19.9 | 38.7±18.7 | 43.5±21.3 | NS |
| F/M ratio | 2.5/1 | 1.9/1 | 3.4/1 | NS |
| Tumour size (cm±s.d.) | 3.7±2.4 | 3.9±2.1 | 3.5±2.3 | NS |
| MACIS (mean±s.d.) | 5.5±1.8 | 5.6±2.0 | 5.5±1.7 | NS |
| Classa | NS | |||
| I | 20 | 11 (55%) | 9 (45%) | |
| II | 6 | 3 (50%) | 3 (50%) | |
| III | 9 | 2 (22%) | 7 (78%) | |
| IV | 3 | 3 (100%) | 0 | |
| Predominant pattern | 0.01 | |||
| Classic | 18 | 5 (28%) | 13 (72%) | |
| Follicular | 15 | 12 (80%) | 3 (20%) | |
| Others | 6 | 3 (50%) | 3(50%) | |
| Poorly differentiated areas | 6 | 4 (67%) | 2 (33%) | NS |
| Local invasion | 13 | 6 (46%) | 7 (54%) | NS |
| Vascular invasion | 9 | 4 (44%) | 5 (56%) | NS |
| Clinical statusa | NS | |||
| Persistent | 9 | 3 (33%) | 6 (67%) | |
| Remission | 29 | 16 (55%) | 13 (45%) | |
PPAR=peroxisome proliferator-activated receptor; PTC=papillary thyroid carcinoma; s.d.=standard deviation; NS=not significant.
Loss of follow-up of one patient.
DISCUSSION
We (Marques et al, 2002) and others (Kroll et al, 2000; Nikiforova et al, 2002; Aldred et al, 2003; Cheung et al, 2003) have detected cases of thyroid tumours, such as FTC, FTA or PTC, that exhibit mild or moderate diffuse PPARγ nuclear staining, even though they are RT–PCR negative for the PAX8-PPARγ fusion gene. The question was then whether such cases present or not overexpression of PPARγ. It is important to discriminate between these two possibilities, because underlying different pathogenic mechanisms may be present. For instance, if PPARγ expression is found to be upregulated in PAX8-PPARγ-negative tumours, it could reflect either a breakpoint between PAX8 and PPARγ in a location outside the primers used in the RT–PCR reaction, or a rearrangement between PPARγ and a non-PAX8 partner, or overexpression of wild-type PPARγ or point mutations in the PPARγ gene. The objectives of the present work were two-fold: 1 – to study, and compare, the relative expression of PPARγ in the normal gland and in benign and malignant diseases of the thyroid; and 2 – to correlate PPARγ immunostaining with clinical and pathological characteristics of patients with thyroid carcinomas of follicular origin. We chose to examine PPARγ expression in thyroid tissues by RT–PCR, interphase FISH, real-time RT–PCR and immunohistochemistry. We first demonstrated that NT tissues express PPARγ both at mRNA and at the protein level. This is in contrast with the findings by Ohta et al (2001), who detected PPARγ mRNA in four of six PTC cell lines and in three of six PTCs, but not in NT tissues or in FTAs. However, our results are in concordance with the recent data of Aldred et al (2003), who have also demonstrated PPARγ expression in seven of seven NT specimens. Interestingly, the mean ratio of PPARγ/GAPDH mRNA obtained by the semiquantitative method of Aldred et al (2003) of 0.79±0.30 is not far from the ratio of 0.68±0.40 obtained by our quantitative method. As the human PPARγ gene gives rise to four mRNAs, PPARγ1–4, that differ at their 5′-end as a consequence of alternate promoter usage and splicing, and these mRNAs code two protein isoforms, PPARγ1 and PPARγ2, which may exert distinct biological effects, we investigated the expression of the different PPARγ transcripts in the thyroid tissues. We were able to show that thyroid cells express all mRNA isoforms, but the proportion of specific variants was similar in normal tissues and in the various types of thyroid tumours studied. However, because we did not perform quantitative RT–PCR, it is possible that some tumour types predominantly express one of the isoforms. To compare PPARγ expression between tumours and NT tissues, we performed quantitative analysis by real-time RT–PCR. Our assay did not distinguish wild-type transcripts from PAX8-PPARγ fusion mRNAs. We observed that tumours negative for the rearrangement expressed lower levels of PPARγ mRNA than NT. This was particularly evident in the PTC cases (n=7) in which the normal adjacent tissue of the same patient was also available for analysis (Figure 2). Upregulation of PPARγ mRNA levels was found in four of the five (80%) translocation-positive tumours (three FTC and two FTA) analysed. However, we detected one PAX8-PPARγ-positive case (FTC) with a PPARγ/GAPDH ratio within the mean of the normal group. Interphase FISH analysis revealed that only a small subset of cells in this case harboured the translocation, which is consistent with the normal expression level of PPARγ as assessed by real-time RT–PCR. Overall, there was a direct correlation between our real-time analysis of PPARγ expression and the immunoreactive protein: strong immunostaining was present only in tumours with upregulated PPARγ mRNA levels and mild or moderate staining was revealed in the remaining tumours, as well as in normal tissues. Notably, the translocation-positive FTC with normal PPARγ/GAPDH ratio showed diffuse and faint nuclear staining. Aldred et al (2003) performed semiquantitative RT–PCR analysis of PPARγ expression in 14 NT tissues and in 19 FTCs and also showed that nontranslocation tumours had underexpression of PPARγ.
A larger number of tissues were examined by immunohistochemistry in order to determine, and compare, the prevalence of PPARγ staining between normal, hyperplastic and neoplastic tissues, and to correlate staining with known prognostic variables of thyroid carcinomas. Compared to NT tissues, staining was significantly (P<0.05) less prevalent in FTCs, PTCs and PDTCs. This trend was also present in FTAs, although not statistically significant (P=0.09). Previous studies have shown that FTCs harbouring the fusion gene, hence strongly reactive with a PPARγ antibody, are somewhat smaller in size (Cheung et al, 2003; Nikiforova et al, 2003), more overtly invasive, and occur at a younger age than tumours without the rearrangement (Nikiforova et al, 2003). In the study of French et al (2003), FTCs with PPARγ rearrangement had vascular invasion and a solid/nested histology more frequently than translocation-negative tumours. We observed that PPARγ-negative FTCs were more likely to be locally invasive, to persist after surgery, to metastasise and to have poorly differentiated areas.
We could not correlate PPARγ staining with any of the prognostic variables analysed in the group of PTCs, except for tumours presenting a predominantly follicular pattern that were more often negative (80%) than classic PTCs (28%; P=0.01).
In summary, we have demonstrated underexpression of PPARγ in PAX8/PPARγ-negative thyroid tumours of follicular origin, and that a further reduction of PPARγ expression is associated with dedifferentiation at later stages of tumour development.
Acknowledgments
We gratefully acknowledge the technical assistance of Ms Teresa Pereira. Ana Rita Marques is a recipient of a PhD fellowship (PRAXIS XXI/BD/21329/99) from Fundação para a Ciência e Tecnologia. This work was supported by Núcleo Regional do Sul Liga Portuguesa Contra O Cancro (Terry Fox) and Fundação para a Ciência e Tecnologia (POCTI/CBO/48922/2002), Portugal.
References
- Aldred MA, Morrison C, Gimm O, Hoang-Vu C, Krause U, Dralle H, Jhiang S, Eng C (2003) Peroxisome proliferator-activated receptor gamma is frequently downregulated in a diversity of sporadic nonmedullary thyroid carcinomas. Oncogene 22: 3412–3416, doi: 10.1038/sj.onc.1206400 [DOI] [PubMed] [Google Scholar]
- Brockman JA, Gupta RA, DuBois RN (1998) Activation of PPARγ leads to inhibition of anchorage-independent growth of human colorectal cancer cells. Gastroenterology 115: 1049–1055 [DOI] [PubMed] [Google Scholar]
- Chang T, Szabo E (2000) Induction of differentiation and apoptosis by ligands of peroxisome proliferator-activated receptor γ in non-small cell lung cancer. Cancer Res 60: 1129–1138 [PubMed] [Google Scholar]
- Chawla A, Lazar MA (1994) Peroxisome proliferator and retinoid signaling pathways co-regulate preadipocyte phenotype and survival. Proc Natl Acad Sci USA 91: 1786–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung L, Messina M, Gill A, Clarkson A, Learoyd D, Delbridge L, Wentworth J, Philips J, Clifton-Bligh R, Robinson BG (2003) Detection of the PAX8-PPARγ fusion oncogene in both follicular thyroid carcinomas and adenomas. J Clin Endocrinol Metab 88: 354–357, doi: 10.1210/jc.2002-021020. [DOI] [PubMed] [Google Scholar]
- DeGroot LJ, Kaplan EL, McCormick M, Straus FH (1990) Natural History, treatment, and course of papillary thyroid carcinoma. J Clin Endocrinol Metab 71: 414–424 [DOI] [PubMed] [Google Scholar]
- Demetri GD, Fletcher CDM, Mueller E, Sarraf P, Naujoks R, Campbell N, Spiegelman B, Singer S (1999) Induction of solid tumor differentiation by the peroxisome proliferator-activated receptor-γ ligand troglitazone in patients with liposarcoma. Proc Natl Acad Sci USA 96: 3951–3956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvergne B, Wahli W (1999) Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev 20: 649–688 [DOI] [PubMed] [Google Scholar]
- Elstner E, Mueller C, Koshizuka K, Williamson EA, Park D, Asou H, Shintaku P, Said JW, Heber D, Koeffler HP (1998) Ligands for peroxisome proliferator-activated receptor-γ and retinoic acid receptor inhibit growth and induce apoptosis of human breast cancer cells in vitro and in BNX mice. Proc Natl Acad Sci USA 95: 8806–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM (1995) 15-Deoxy-Δ12,14 prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell 83: 803–812 [DOI] [PubMed] [Google Scholar]
- French CA, Alexander EK, Cibas ES, Nose V, Laguette J, Faquin W, Garber J, Moore Jr F, Fletcher JA, Larsen PR, Kroll TG (2003) Genetic and biological subgroups of low-stage follicular thyroid cancer. Am J Pathol 162: 1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan YF, Zhang YH, Breyer RM, Davis L, Breyer MD 1999. Expression of peroxisome proliferator-activated receptor gamma (PPAR gamma) in human transitional bladder cancer and its role in inducing cell death. Neoplasia 1: 330–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase T, Yoshimura R, Mitsuhashi M, Segawa Y, Kawashito Y, Wada S, Nakatani T, Sano H (2002) Expression of peroxisome proliferator-activated receptors in human testicular cancer and growth inhibition by its agonists. Urology 60: 542–547 [DOI] [PubMed] [Google Scholar]
- Hay ID, Bergstralh EJ, Goellner JR, Ebersold JR, Grant, CS (1993) Predicting outcome in papillary thyroid carcinoma: development of a reliable prognostic scoring system in a cohort of 1779 patients surgically treated at one institution during 1940 through 1989. Surgery 114: 1050–1058 [PubMed] [Google Scholar]
- Inoue K, Kawashito Y, Tsubouchi Y, Kohno M, Yoshimura R, Yoshikawa T, Sano H 2001. Expression of peroxisome proliferator-activated receptor γ in renal cell carcinoma and growth inhibition by its agonists. Biochem Biophys Res Commun 287: 727–732 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Miyazaki Y, Shinomura Y, Kondo S, Kanayama S, Matsuzawa Y (1999) Peroxisome proliferator-activated receptor gamma induces growth arrest and differentiation markers of human colon cancer cells. Jpn J Cancer Res 90: 75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehman JM (1995) A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell 83: 813–819 [DOI] [PubMed] [Google Scholar]
- Kroll TG, Sarraf P, Pecciarini L, Chen C, Mueller E, Spiegelman BM, Fletcher JA (2000) PAX8-PPARγ1 fusion oncogene in human thyroid carcinoma. Science 289: 1357–1360 [DOI] [PubMed] [Google Scholar]
- Kubota T, Koshizika K, Williamson EA, Asou H, Said JW, Holden S, Miyoshi I, Koeffler HP (1998) Ligand for peroxisome proliferator-activated receptor γ (troglitazone) has potent antitumor effect against human prostate cancer both in vitro and in vivo. Cancer Res 58: 3344–3352 [PubMed] [Google Scholar]
- Lazar V, Bidart JM, Caillou B, Mahé C, Lacroix L, Filetti S, Schumberger M (1999) Expression of the Na+/I− symporter gene in human thyroid tumors: a comparison study with other thyroid-specific genes. J Clin Endocrinol Metab 84: 3228–3234 [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer AS (1995) An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPAR γ). J Biol Chem 270: 12953–12956 [DOI] [PubMed] [Google Scholar]
- Marques AR, Espadinha C, Catarino AL, Moniz S, Pereira T, Sobrinho LG, Leite V (2002) Expression of PAX8-PPARγ1 rearrangements in both follicular thyroid carcinomas and adenomas. J Clin Endocrinol Metab 87: 3947–3952 [DOI] [PubMed] [Google Scholar]
- Martelli ML, Iuliano R, Le Pera I, Sama I, Monaco C, Cammarota S, Kroll T, Chiariotti L, Santoro M, Fusco A (2002) Inhibitory effects of peroxisome proliferator-activated receptor γ on thyroid carcinoma cell growth. J Clin Endocrinol Metab 87: 4728–4735, doi: 10.1210/jc.2001-012054 [DOI] [PubMed] [Google Scholar]
- Martin G, Schoonjans K, Staels B, Auwerx J (1998) PPAR-gamma activators improve glucose homeostasis by stimulating fatty acid uptake in the adipocytes. Atherosclerosis 137: S75–S80 [DOI] [PubMed] [Google Scholar]
- Motomura W, Okumur T, Takahashi N, Obara T, Kohgo Y (2000) Activation of peroxisome proliferator-activated receptor gamma by troglitazone inhibits cell growth through the increase of p27 Kip1 in human pancreatic carcinoma cells. Cancer Res 60: 5558–5564 [PubMed] [Google Scholar]
- Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, Fletcher C, Singer S, Spiegeman BM (1998) Terminal differentiation of human breast cancer through PPAR-gamma. Mol Cell 1: 465–470 [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Biddinger PW, Caudill CM, Kroll TG, Nikiforov YE (2002) PAX8-PPARγ rearrangement in thyroid tumors – RT–PCR and immunohistochemical analysis. Am J Surg Pathol 26: 1016–1023, doi: 10.1097/01.PAS.0000021228.30983.6A [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn II GW, Tallini G, Kroll TG, Nikiforov YE (2003) RAS point mutations and PAX8-PPAR gamma rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab 88: 2318–2326, doi: 10.1210/jc.2002-021907 [DOI] [PubMed] [Google Scholar]
- Ohta K, Endo T, Haraguchi K, Hershman JM, Onaya T (2001) Ligands for peroxisome proliferator-activated receptor γ inhibit growth and induce apoptosis of human papillary thyroid carcinoma cells. J Clin Endocrinol Metab 86: 2170–2177 [DOI] [PubMed] [Google Scholar]
- Rosen ED, Spiegelman BM (2001) PPARγ: a nuclear regulator of metabolism, differentiation and cell growth. J Biol Chem 276: 37731–37734, doi: 10.1074/jbc.R100034200 [DOI] [PubMed] [Google Scholar]
- Sarraf P, Mueller E, Jones D, King FJ, DeAngelo DJ, Partridge JB, Holden SA, Chen LB, Singer S, Fletcher C, Spiegelman BM (1998) Differentiation and reversal of malignant changes in colon cancer through PPARγ. Nat Med 4: 1046–1052 [DOI] [PubMed] [Google Scholar]
- Sarraf P, Mueller E, Smith WM, Wright HM, Kum JB, Aaltonen LA, de la Chapelle A, Spiegelman BM, Eng C (1999) Loss-of-function mutations in PPARγ associated with human colon cancer. Cell 3: 799–804 [DOI] [PubMed] [Google Scholar]
- Sato H, Ishihara S, Kawashima K, Moriyama N, Suetsugu H, Kazumori H, Okuyama T, Rumi MA, Fukuda R, Nagasue N, Kinoshita Y (2000) Expression of peroxisome proliferator-activated receptor (PPAR) gamma in gastric cancer and inhibitory effects of PPAR gamma agonists. Br J Cancer 83: 1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugg SL, Ezzat S, Rosen IB, Freeman JL, Asa SL (1998) Distinct multiple RET/PTC gene rearrangements in multifocal papillary thyroid neoplasia. J Clin Endocrinol Metab 83: 4116–4122 [DOI] [PubMed] [Google Scholar]
- Sundvold H, Lien S (2001) Identification of a novel peroxisome proliferator-activated receptor (PPAR) gamma promoter in man and transactivation by the nuclear receptor ROR alpha 1. Biochem Biophys Res Commun 287: 383–390 [DOI] [PubMed] [Google Scholar]
- Terashita Y, Sasaki H, Haruki N, Nishiwaki T, Ishiguro H, Shibata Y, Kudo J, Konishi S, Kato J, Koyama H, Kimura M, Sato A, Shinoda N, Kuwabara Y, Fujii Y (2002) Decreased peroxisome proliferator-activated receptor gamma gene expression is correlated with poor prognosis in patients with esophageal cancer. Jpn J Clin Oncol 32: 238–243 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Buclavari AI, Spiegelman BM (1994) mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 8: 1224–1234 [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Singer S, Forman BM, Sarraf P, Fletcher JA, Fletcher CDM, Brun RP, Mueller E, Altiok S, Oppenheim H, Evans RM, Spiegelman BM (1997) Terminal differentiation of human liposarcoma cells induced by ligands for peroxisome proliferator-activated receptor γ and the retinoid X receptor. Proc Natl Acad Sci USA 94: 237–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda M, Takagi H, Noriguchi N, Kakizaki S, Sato H, Takayama H, Mori M (2002) A ligand for peroxisome proliferator activated receptor γ inhibits cell growth and induces apoptosis in human liver cancer cells. Gut 50: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]