Abstract
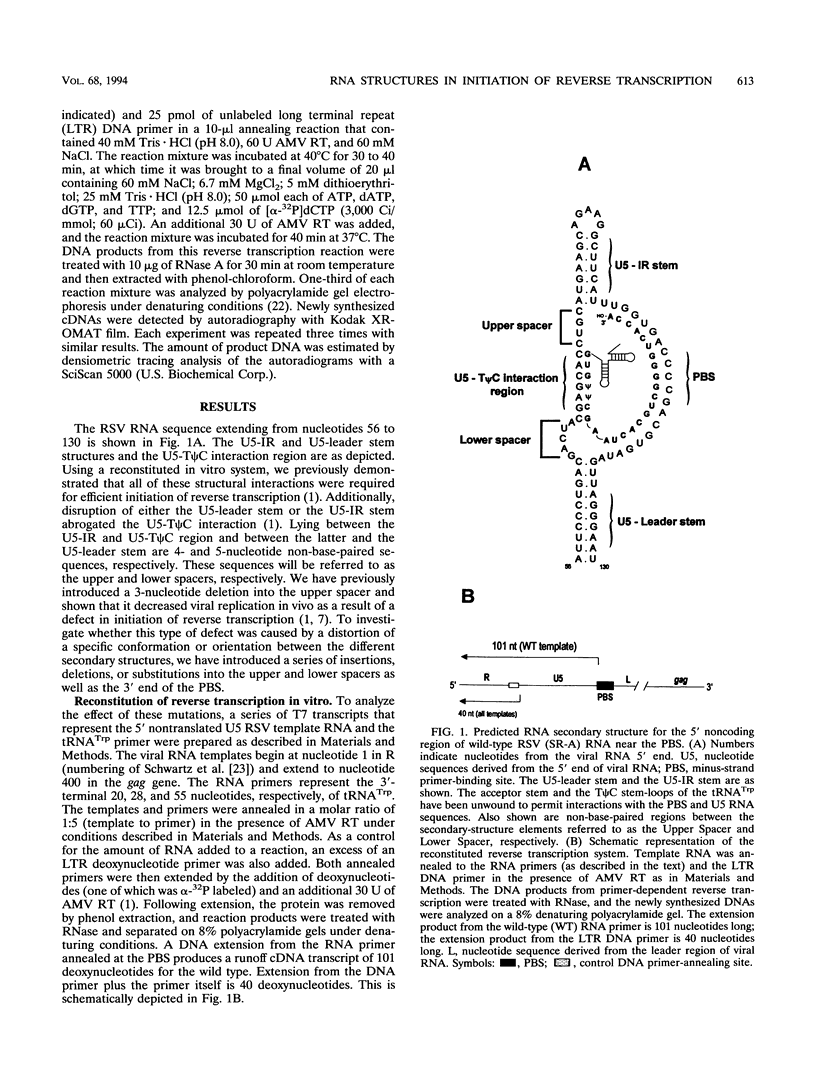
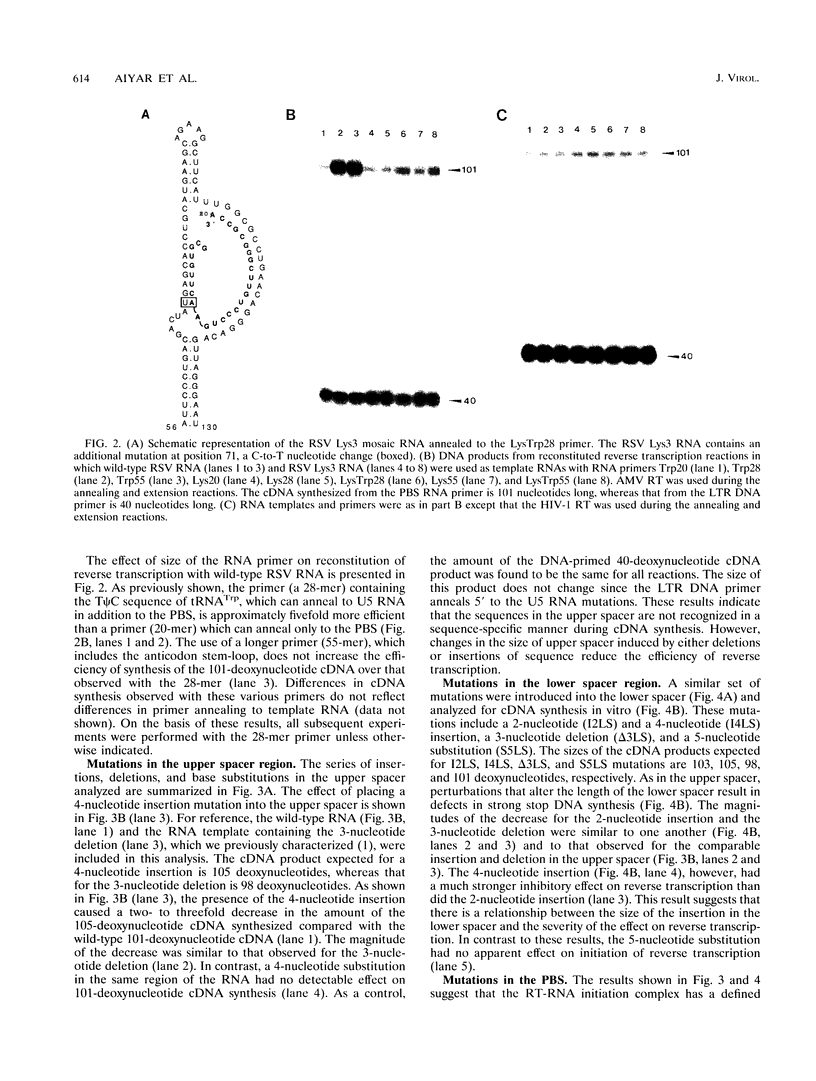
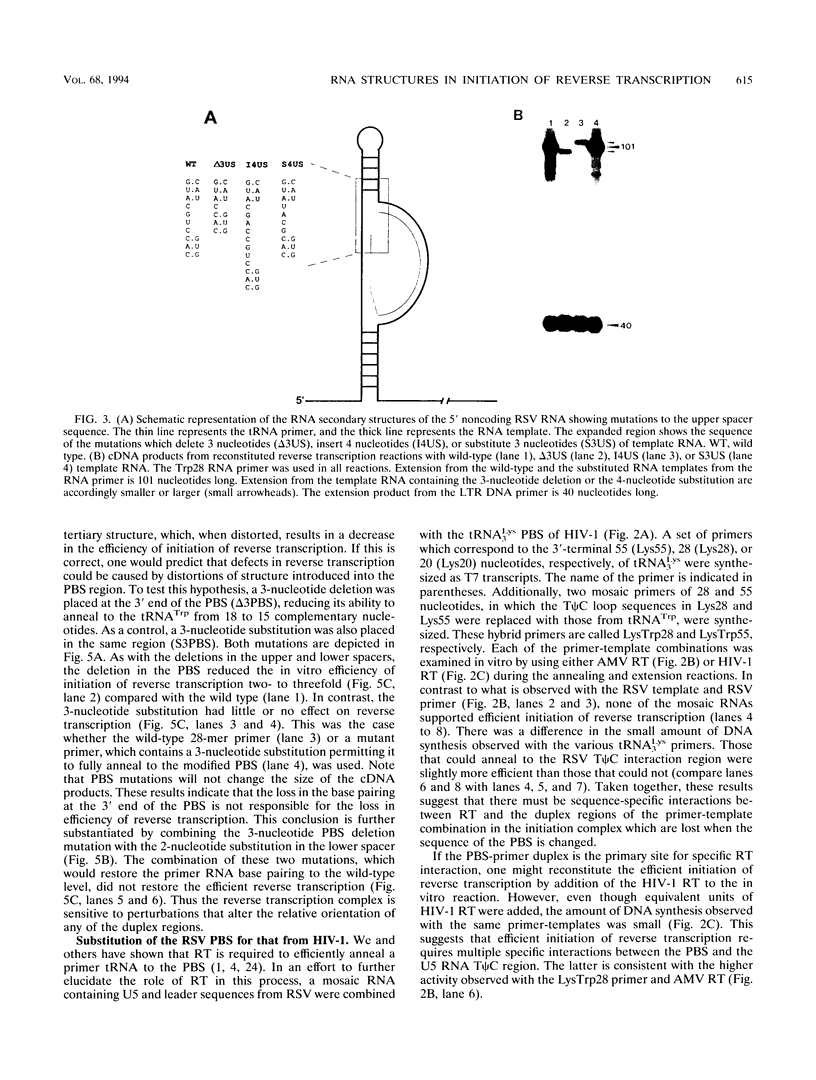
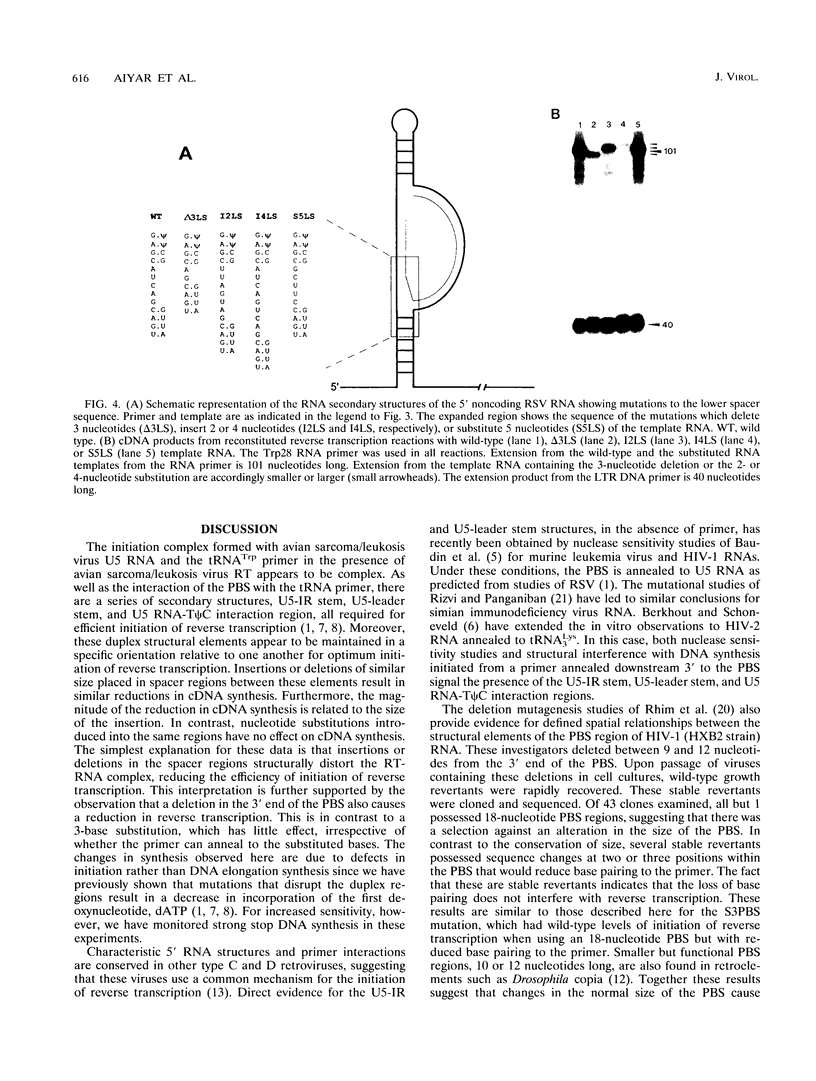
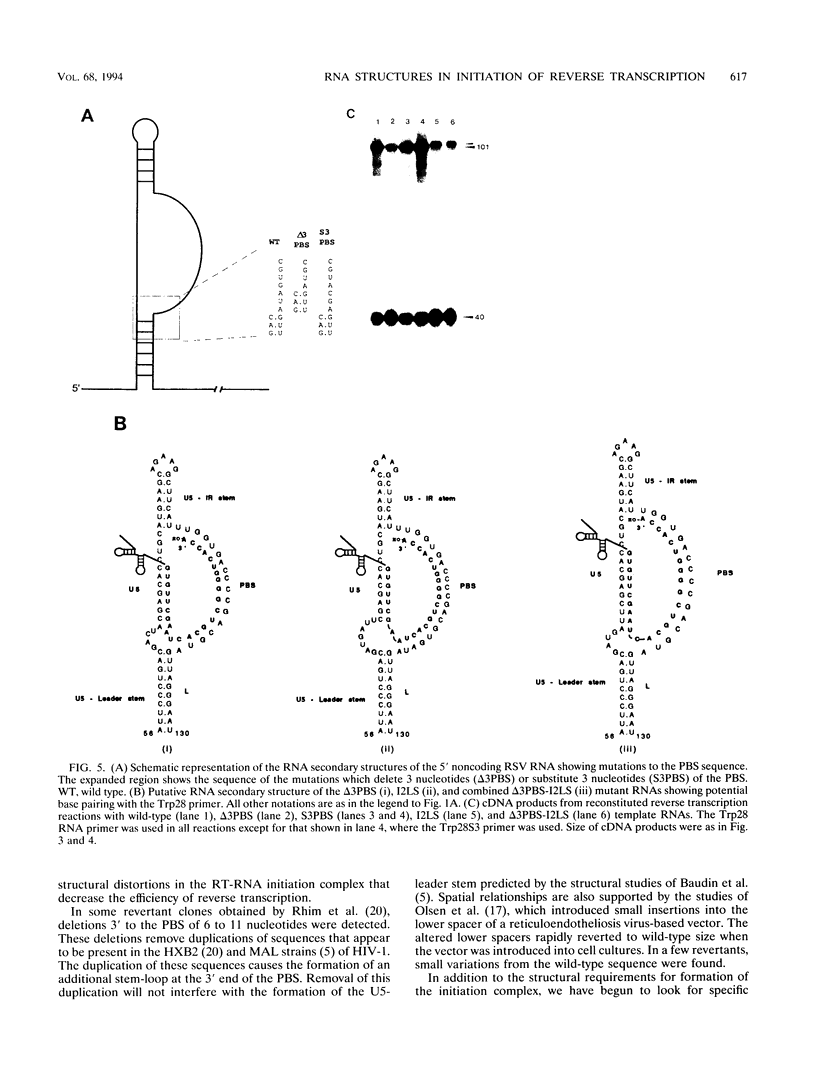
The 5' end of avian retrovirus RNA near the primer-binding site (PBS) forms two secondary structures, the U5-inverted repeat (U5-IR) and the U5-leader stems, and contains a 7-nucleotide sequence that anneals to the T psi C loop of the tRNA(Trp) primer. Mutations that disrupt any of these base pair interactions cause defects in initiation of reverse transcription both in vivo and in vitro (D. Cobrinik, A. Aiyar, Z. Ge, M. Katzman, H. Huang, and J. Leis, J. Virol. 65:3864-3872, 1991; A. Aiyar, D. Cobrinik, Z. Ge, H.-J. Kung, and J. Leis, J. Virol. 66:2464-2472, 1992). We have now examined the effect of perturbing the non-base-paired intervening "spacer" sequences between these secondary-structure elements. Small deletions or insertions in these intervening sequences decreased initiation of reverse transcription in vitro. In contrast, base substitutions, which maintain the spacing distances between the structures, had no detectable effect. Additionally, a small deletion at the 3' end of the PBS caused a significant decrease in initiation of reverse transcription whereas substitution mutations again had no effect. Together, these results indicate that reverse transcriptase forms a complex in which the different structural elements are maintained in a specific orientation that is required for efficient initiation of reverse transcription. Specific sequence recognition of the duplex structures by reverse transcriptase is also required since mosaic RNAs that combine the human immunodeficiency virus type 1 PBS with avian sequences is not efficiently utilized for reverse transcription even though the primer used can anneal to the substituted PBS.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiyar A., Cobrinik D., Ge Z., Kung H. J., Leis J. Interaction between retroviral U5 RNA and the T psi C loop of the tRNA(Trp) primer is required for efficient initiation of reverse transcription. J Virol. 1992 Apr;66(4):2464–2472. doi: 10.1128/jvi.66.4.2464-2472.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A., Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. Biotechniques. 1993 Mar;14(3):366–369. [PubMed] [Google Scholar]
- Barat C., Le Grice S. F., Darlix J. L. Interaction of HIV-1 reverse transcriptase with a synthetic form of its replication primer, tRNA(Lys,3). Nucleic Acids Res. 1991 Feb 25;19(4):751–757. doi: 10.1093/nar/19.4.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin F., Marquet R., Isel C., Darlix J. L., Ehresmann B., Ehresmann C. Functional sites in the 5' region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993 Jan 20;229(2):382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Schoneveld I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993 Mar 11;21(5):1171–1178. doi: 10.1093/nar/21.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Aiyar A., Ge Z., Katzman M., Huang H., Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J Virol. 1991 Jul;65(7):3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D., Soskey L., Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol. 1988 Oct;62(10):3622–3630. doi: 10.1128/jvi.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. C., Dahlberg J. E. Structural features required for the binding of tRNATrp to avian myeloblastosis virus reverse transcriptase. Nucleic Acids Res. 1983 Jul 25;11(14):4823–4833. doi: 10.1093/nar/11.14.4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Mak J., Ladha A., Cohen E., Klein M., Rovinski B., Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993 Jun;67(6):3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Ando Y., Shiba T. Unusual priming mechanism of RNA-directed DNA synthesis in copia retrovirus-like particles of Drosophila. 1986 Oct 30-Nov 5Nature. 323(6091):824–826. doi: 10.1038/323824a0. [DOI] [PubMed] [Google Scholar]
- Levin J. G., Seidman J. G. Selective packaging of host tRNA's by murine leukemia virus particles does not require genomic RNA. J Virol. 1979 Jan;29(1):328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. E., Goff S. P. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: effects on RNA packaging and reverse transcription. J Virol. 1989 Jan;63(1):319–327. doi: 10.1128/jvi.63.1.319-327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson P., Temin H. M., Dornburg R. Unusually high frequency of reconstitution of long terminal repeats in U3-minus retrovirus vectors by DNA recombination or gene conversion. J Virol. 1992 Mar;66(3):1336–1343. doi: 10.1128/jvi.66.3.1336-1343.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. G., Hu J. Reverse transcriptase as the major determinant for selective packaging of tRNA's into Avian sarcoma virus particles. J Virol. 1980 Dec;36(3):692–700. doi: 10.1128/jvi.36.3.692-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats A. C., Sarih L., Gabus C., Litvak S., Keith G., Darlix J. L. Small finger protein of avian and murine retroviruses has nucleic acid annealing activity and positions the replication primer tRNA onto genomic RNA. EMBO J. 1988 Jun;7(6):1777–1783. doi: 10.1002/j.1460-2075.1988.tb03008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim H., Park J., Morrow C. D. Deletions in the tRNA(Lys) primer-binding site of human immunodeficiency virus type 1 identify essential regions for reverse transcription. J Virol. 1991 Sep;65(9):4555–4564. doi: 10.1128/jvi.65.9.4555-4564.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi T. A., Panganiban A. T. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J Virol. 1993 May;67(5):2681–2688. doi: 10.1128/jvi.67.5.2681-2688.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D. E., Tizard R., Gilbert W. Nucleotide sequence of Rous sarcoma virus. Cell. 1983 Mar;32(3):853–869. doi: 10.1016/0092-8674(83)90071-5. [DOI] [PubMed] [Google Scholar]
- Weiss S., König B., Müller H. J., Seidel H., Goody R. S. Synthetic human tRNA(UUULys3) and natural bovine tRNA(UUULys3) interact with HIV-1 reverse transcriptase and serve as specific primers for retroviral cDNA synthesis. Gene. 1992 Feb 15;111(2):183–197. doi: 10.1016/0378-1119(92)90686-j. [DOI] [PubMed] [Google Scholar]