Abstract
Influenza virus polymerase complex is a heterotrimer consisting of polymerase basic protein 1 (PB1), polymerase basic protein 2 (PB2), and polymerase acidic protein (PA). Of these, only PB1, which has been implicated in RNA chain elongation, possesses the four conserved motifs (motifs I, II, III, and IV) and the four invariant amino acids (one in each motif) found among all viral RNA-dependent RNA or RNA-dependent DNA polymerases. We have modified an assay system developed by Huang et al. (T.-J. Huang, P. Palese, and M. Krystal, J. Virol. 64:5669-5673, 1990) to reconstitute the functional polymerase activity in vivo. Using this assay, we have examined the requirement of each of these motifs of PB1 in polymerase activity. We find that each of these invariant amino acids is critical for PB1 activity and that mutation in any one of these residues renders the protein nonfunctional. We also find that in motif III, which contains the SSDD sequence, the signature sequence of influenza virus RNA polymerase, SDD is essentially invariant and cannot accommodate sequences found in other RNA viral polymerases. However, conserved changes in the flanking sequences of SDD can be partially tolerated. These results provide the experimental evidence that influenza virus PB1 possesses a similar polymerase module as has been proposed for other RNA viruses and that the core SDD sequence of influenza virus PB1 represents a sequence variant of the GDN in negative-stranded nonsegmented RNA viruses, GDD in positive-stranded RNA virus and double-stranded RNA viruses, or MDD in retroviruses.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akkina R. K., Chambers T. M., Londo D. R., Nayak D. P. Intracellular localization of the viral polymerase proteins in cells infected with influenza virus and cells expressing PB1 protein from cloned cDNA. J Virol. 1987 Jul;61(7):2217–2224. doi: 10.1128/jvi.61.7.2217-2224.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkina R. K., Richardson J. C., Aguilera M. C., Yang C. M. Heterogeneous forms of polymerase proteins exist in influenza A virus-infected cells. Virus Res. 1991 Mar;19(1):17–30. doi: 10.1016/0168-1702(91)90091-9. [DOI] [PubMed] [Google Scholar]
- Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988 Nov 11;16(21):9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton A. R., Krug R. M. Synthesis of the templates for influenza virion RNA replication in vitro. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4682–4686. doi: 10.1073/pnas.81.15.4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton A. R., Krug R. M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5' capped end. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P. L., Ferris A. L., Hughes S. H. Cassette mutagenesis of the reverse transcriptase of human immunodeficiency virus type 1. J Virol. 1992 Feb;66(2):1031–1039. doi: 10.1128/jvi.66.2.1031-1039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J., Ulmanen I., Krug R. M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell. 1983 Sep;34(2):609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- Castrucci M. R., Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol. 1993 Feb;67(2):759–764. doi: 10.1128/jvi.67.2.759-764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C. N., MacWilliams H. Regulation of the self-renewal probability in Hydra stem cell clones. Proc Natl Acad Sci U S A. 1978 Feb;75(2):886–890. doi: 10.1073/pnas.75.2.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami M., Luytjes W., Krystal M., Palese P. Introduction of site-specific mutations into the genome of influenza virus. Proc Natl Acad Sci U S A. 1990 May;87(10):3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner P. L., Gadek T. R., Holm M., Roman R., Chan H. W., Wenz M., Northrop J. P., Ringold G. M., Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Earl P. L., Moss B. Use of a hybrid vaccinia virus-T7 RNA polymerase system for expression of target genes. Mol Cell Biol. 1987 Jul;7(7):2538–2544. doi: 10.1128/mcb.7.7.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman C. M., Moffat L. F., Howard B. H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982 Sep;2(9):1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyward J. T., Klimas R. A., Stapp M. D., Obijeski J. F. The rapid concentration and purification of influenza virus from allantoic fluid. Arch Virol. 1977;55(1-2):107–119. doi: 10.1007/BF01314484. [DOI] [PubMed] [Google Scholar]
- Hizi A., Barber A., Hughes S. H. Effects of small insertions on the RNA-dependent DNA polymerase activity of HIV-1 reverse transcriptase. Virology. 1989 May;170(1):326–329. doi: 10.1016/0042-6822(89)90389-9. [DOI] [PubMed] [Google Scholar]
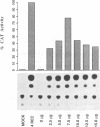
- Huang T. S., Palese P., Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990 Nov;64(11):5669–5673. doi: 10.1128/jvi.64.11.5669-5673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski S. A., Luo M., Morrow C. D. Enzymatic activity of poliovirus RNA polymerase mutants with single amino acid changes in the conserved YGDD amino acid motif. J Virol. 1991 Sep;65(9):4565–4572. doi: 10.1128/jvi.65.9.4565-4572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski S. A., Morrow C. D. Enzymatic activity of poliovirus RNA polymerases with mutations at the tyrosine residue of the conserved YGDD motif: isolation and characterization of polioviruses containing RNA polymerases with FGDD and MGDD sequences. J Virol. 1993 Jan;67(1):373–381. doi: 10.1128/jvi.67.1.373-381.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamer G., Argos P. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 1984 Sep 25;12(18):7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemdirim S., Palefsky J., Briedis D. J. Influenza B virus PB1 protein; nucleotide sequence of the genome RNA segment predicts a high degree of structural homology with the corresponding influenza A virus polymerase protein. Virology. 1986 Jul 15;152(1):126–135. doi: 10.1016/0042-6822(86)90378-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Tuchiya K., Nagata K., Ishihama A. Reconstitution of influenza virus RNA polymerase from three subunits expressed using recombinant baculovirus system. Virus Res. 1992 Mar;22(3):235–245. doi: 10.1016/0168-1702(92)90055-e. [DOI] [PubMed] [Google Scholar]
- Krug R. M., Ueda M., Palese P. Temperature-sensitive mutants of influenza WSN virus defective in virus-specific RNA synthesis. J Virol. 1975 Oct;16(4):790–796. doi: 10.1128/jvi.16.4.790-796.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B. A., Purifoy D. J., Powell K. L., Darby G. Site-specific mutagenesis of AIDS virus reverse transcriptase. 1987 Jun 25-Jul 1Nature. 327(6124):716–717. doi: 10.1038/327716a0. [DOI] [PubMed] [Google Scholar]
- Li X., Palese P. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J Virol. 1992 Jul;66(7):4331–4338. doi: 10.1128/jvi.66.7.4331-4338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Bergmann M., Garcia-Sastre A., Palese P. Mechanism of attenuation of a chimeric influenza A/B transfectant virus. J Virol. 1992 Aug;66(8):4679–4685. doi: 10.1128/jvi.66.8.4679-4685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Krystal M., Enami M., Parvin J. D., Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989 Dec 22;59(6):1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- Malone R. W., Felgner P. L., Verma I. M. Cationic liposome-mediated RNA transfection. Proc Natl Acad Sci U S A. 1989 Aug;86(16):6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaigawa J., Nayak D. P. Two signals mediate nuclear localization of influenza virus (A/WSN/33) polymerase basic protein 2. J Virol. 1991 Jan;65(1):245–253. doi: 10.1128/jvi.65.1.245-253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T., Subbarao E. K., Enami M., Murphy B. R., Palese P. An influenza A virus containing influenza B virus 5' and 3' noncoding regions on the neuraminidase gene is attenuated in mice. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5177–5181. doi: 10.1073/pnas.88.12.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath S. T., Nayak D. P. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1). Mol Cell Biol. 1990 Aug;10(8):4139–4145. doi: 10.1128/mcb.10.8.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik A. K., Wertz G. W. Cells that express all five proteins of vesicular stomatitis virus from cloned cDNAs support replication, assembly, and budding of defective interfering particles. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1379–1383. doi: 10.1073/pnas.88.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik A. K., Wertz G. W. Replication and amplification of defective interfering particle RNAs of vesicular stomatitis virus in cells expressing viral proteins from vectors containing cloned cDNAs. J Virol. 1990 Jun;64(6):2948–2957. doi: 10.1128/jvi.64.6.2948-2957.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Blumberg B. M., Bougueleret L., Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990 May;71(Pt 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Poch O., Sauvaget I., Delarue M., Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989 Dec 1;8(12):3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V. R., Goff S. P. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci U S A. 1989 May;86(9):3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello V. R., Baltimore D. Molecular cloning of poliovirus cDNA and determination of the complete nucleotide sequence of the viral genome. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4887–4891. doi: 10.1073/pnas.78.8.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Ribas J. C., Wickner R. B. RNA-dependent RNA polymerase consensus sequence of the L-A double-stranded RNA virus: definition of essential domains. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2185–2189. doi: 10.1073/pnas.89.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro G. I., Krug R. M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988 Jul;62(7):2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian N., Nayak D. P. Sequence analysis of the polymerase 1 gene and the secondary structure prediction of polymerase 1 protein of human influenza virus A/WSN/33. J Virol. 1982 Oct;44(1):321–329. doi: 10.1128/jvi.44.1.321-329.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleat D. E., Banerjee A. K. Transcriptional activity and mutational analysis of recombinant vesicular stomatitis virus RNA polymerase. J Virol. 1993 Mar;67(3):1334–1339. doi: 10.1128/jvi.67.3.1334-1339.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. L., Levin J. Z., Palese P., Moss B. Synthesis and cellular location of the ten influenza polypeptides individually expressed by recombinant vaccinia viruses. Virology. 1987 Oct;160(2):336–345. doi: 10.1016/0042-6822(87)90004-3. [DOI] [PubMed] [Google Scholar]
- Szewczyk B., Laver W. G., Summers D. F. Purification, thioredoxin renaturation, and reconstituted activity of the three subunits of the influenza A virus RNA polymerase. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7907–7911. doi: 10.1073/pnas.85.21.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Broni B. A., Krug R. M. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Broni B., Krug R. M. Influenza virus temperature-sensitive cap (m7GpppNm)-dependent endonuclease. J Virol. 1983 Jan;45(1):27–35. doi: 10.1128/jvi.45.1.27-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield J. K., Jablonski S. A., Morrow C. D. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J Virol. 1992 Nov;66(11):6806–6812. doi: 10.1128/jvi.66.11.6806-6812.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Ogasawara N., Yoshikawa H., Ishihama A., Nagata K. In vivo analysis of the promoter structure of the influenza virus RNA genome using a transfection system with an engineered RNA. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5369–5373. doi: 10.1073/pnas.88.12.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M., Krystal M., Palese P. Comparison of the three large polymerase proteins of influenza A, B, and C viruses. Virology. 1989 Aug;171(2):458–466. doi: 10.1016/0042-6822(89)90615-6. [DOI] [PubMed] [Google Scholar]