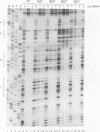
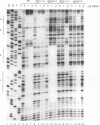
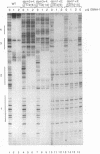
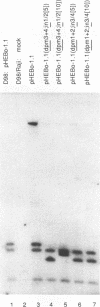
Abstract
The Epstein-Barr virus (EBV) latent origin of DNA replication (oriP) is composed of two elements that contain binding sites for the sole viral gene product required for latent cycle replication, EBNA-1. One of these elements, region I, functions as an EBNA-1-dependent enhancer for RNA polymerase II-transcribed genes, may play a role in plasmid segregation, and is required for origin function in B cells latently infected with EBV. The second element, region II, contains or is very near the site of initiation of DNA replication. A genetic approach was taken to determine the contribution of the EBNA-1 binding sites in oriP to origin function. Although region I is required for the transient replication of plasmids bearing region II in EBV-infected B cells, a plasmid lacking region I but containing region II, was observed to replicate transiently in both D98/Raji and HeLa cells expressing EBNA-1. Thus, binding of EBNA-1 to region I is not absolutely required for the molecular events that lead to initiation of DNA replication at region II. Site-directed mutagenesis of the four EBNA-1-binding sites in region II, individually and in various combinations, demonstrated that only two EBNA-1-binding sites are required for region II function. The results obtained with these mutants, together with the analysis of the replicative ability of plasmids containing insertions between EBNA-1-binding sites, suggested that the spatial relationship of the two sites is critical. Mutants that contain only two EBNA-1-binding sites separated by 26 to 31 bp in region II were not maintained as plasmids over many cell generations and were greatly reduced in their ability to replicate transiently in D98/Raji cells. The EBNA-1-induced bending or untwisting of the DNA in EBNA-1-binding sites 1 and 4 in region II did not, however, demonstrate this spatial constraint. It may be concluded from these results that specific protein-protein interactions between EBNA-1 and/or between EBNA-1 and a cellular protein(s) are required for origin function.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. Replication of latent Epstein-Barr virus genomes in Raji cells. J Virol. 1987 May;61(5):1743–1746. doi: 10.1128/jvi.61.5.1743-1746.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambinder R. F., Mullen M. A., Chang Y. N., Hayward G. S., Hayward S. D. Functional domains of Epstein-Barr virus nuclear antigen EBNA-1. J Virol. 1991 Mar;65(3):1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambinder R. F., Shah W. A., Rawlins D. R., Hayward G. S., Hayward S. D. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J Virol. 1990 May;64(5):2369–2379. doi: 10.1128/jvi.64.5.2369-2379.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Dean F. B., Bullock P. A., Hurwitz J. Binding and unwinding--how T antigen engages the SV40 origin of DNA replication. Cell. 1990 Jan 26;60(2):181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- Borowiec J. A., Hurwitz J. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 1988 Oct;7(10):3149–3158. doi: 10.1002/j.1460-2075.1988.tb03182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowiec J. A., Zhang L., Sasse-Dwight S., Gralla J. D. DNA supercoiling promotes formation of a bent repression loop in lac DNA. J Mol Biol. 1987 Jul 5;196(1):101–111. doi: 10.1016/0022-2836(87)90513-4. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Chittenden T., Lupton S., Levine A. J. Functional limits of oriP, the Epstein-Barr virus plasmid origin of replication. J Virol. 1989 Jul;63(7):3016–3025. doi: 10.1128/jvi.63.7.3016-3025.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar V., Schildkraut C. L. Role of EBNA-1 in arresting replication forks at the Epstein-Barr virus oriP family of tandem repeats. Mol Cell Biol. 1991 Dec;11(12):6268–6278. doi: 10.1128/mcb.11.12.6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornreiter I., Höss A., Arthur A. K., Fanning E. SV40 T antigen binds directly to the large subunit of purified DNA polymerase alpha. EMBO J. 1990 Oct;9(10):3329–3336. doi: 10.1002/j.1460-2075.1990.tb07533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. EBNA1 distorts oriP, the Epstein-Barr virus latent replication origin. J Virol. 1992 Mar;66(3):1786–1790. doi: 10.1128/jvi.66.3.1786-1790.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. Epstein-Barr nuclear antigen 1 mediates a DNA loop within the latent replication origin of Epstein-Barr virus. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10875–10879. doi: 10.1073/pnas.88.23.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappier L., O'Donnell M. Overproduction, purification, and characterization of EBNA1, the origin binding protein of Epstein-Barr virus. J Biol Chem. 1991 Apr 25;266(12):7819–7826. [PubMed] [Google Scholar]
- Gahn T. A., Schildkraut C. L. The Epstein-Barr virus origin of plasmid replication, oriP, contains both the initiation and termination sites of DNA replication. Cell. 1989 Aug 11;58(3):527–535. doi: 10.1016/0092-8674(89)90433-9. [DOI] [PubMed] [Google Scholar]
- Glaser R., Nonoyama M. Host cell regulation of induction of Epstein-Barr virus. J Virol. 1974 Jul;14(1):174–176. doi: 10.1128/jvi.14.1.174-176.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Rapid "footprinting" on supercoiled DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3078–3081. doi: 10.1073/pnas.82.10.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan E. A., Summers W. P., Dowling S., Shedd D., Gradoville L., Miller G. Two Epstein-Barr viral nuclear neoantigens distinguished by gene transfer, serology, and chromosome binding. Proc Natl Acad Sci U S A. 1983 Dec;80(24):7650–7653. doi: 10.1073/pnas.80.24.7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussander E., Adams A. Electron microscopic evidence for replication of circular Epstein-Barr virus genomes in latently infected Raji cells. J Virol. 1984 Nov;52(2):549–556. doi: 10.1128/jvi.52.2.549-556.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampar B., Tanaka A., Nonoyama M., Derge J. G. Replication of the resident repressed Epstein-Barr virus genome during the early S phase (S-1 period) of nonproducer Raji cells. Proc Natl Acad Sci U S A. 1974 Mar;71(3):631–633. doi: 10.1073/pnas.71.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A., Young B. D., Griffin B. E. Random association of Epstein-Barr virus genomes with host cell metaphase chromosomes in Burkitt's lymphoma-derived cell lines. J Virol. 1985 Oct;56(1):328–332. doi: 10.1128/jvi.56.1.328-332.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing J. C., Levine A. J. The Epstein-Barr virus nuclear antigen (BamHI K antigen) is a single-stranded DNA binding phosphoprotein. Virology. 1985 Aug;145(1):105–116. doi: 10.1016/0042-6822(85)90205-3. [DOI] [PubMed] [Google Scholar]
- Hearing J. C., Nicolas J. C., Levine A. J. Identification of Epstein-Barr virus sequences that encode a nuclear antigen expressed in latently infected lymphocytes. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4373–4377. doi: 10.1073/pnas.81.14.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing J., Mülhaupt Y., Harper S. Interaction of Epstein-Barr virus nuclear antigen 1 with the viral latent origin of replication. J Virol. 1992 Feb;66(2):694–705. doi: 10.1128/jvi.66.2.694-705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy K., Kieff E. One of two Epstein-Barr virus nuclear antigens contains a glycine-alanine copolymer domain. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5665–5669. doi: 10.1073/pnas.80.18.5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Karlin S., Blaisdell B. E. A model for the development of the tandem repeat units in the EBV ori-P region and a discussion of their possible function. J Mol Evol. 1987;25(3):215–229. doi: 10.1007/BF02100015. [DOI] [PubMed] [Google Scholar]
- Kimball A. S., Milman G., Tullius T. D. High-resolution footprints of the DNA-binding domain of Epstein-Barr virus nuclear antigen 1. Mol Cell Biol. 1989 Jun;9(6):2738–2742. doi: 10.1128/mcb.9.6.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A., Schwedes J. F., Tegtmeyer P. Herpes simplex virus origin-binding protein (UL9) loops and distorts the viral replication origin. J Virol. 1991 Jun;65(6):3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J., Haase S. B., Calos M. P. Isolation of human sequences that replicate autonomously in human cells. Mol Cell Biol. 1989 Mar;9(3):1026–1033. doi: 10.1128/mcb.9.3.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. K., Horikoshi M., Roeder R. G. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991 Dec 20;67(6):1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Sung N. S., Pesano R. L., Sexton C. J., Hutchison C., 3rd, Pagano J. S. Plasmid origin of replication of herpesvirus papio: DNA sequence and enhancer function. J Virol. 1990 Jun;64(6):2876–2883. doi: 10.1128/jvi.64.6.2876-2883.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton S., Levine A. J. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol Cell Biol. 1985 Oct;5(10):2533–2542. doi: 10.1128/mcb.5.10.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mecsas J., Sugden B. Replication of plasmids derived from bovine papilloma virus type 1 and Epstein-Barr virus in cells in culture. Annu Rev Cell Biol. 1987;3:87–108. doi: 10.1146/annurev.cb.03.110187.000511. [DOI] [PubMed] [Google Scholar]
- Melendy T., Stillman B. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J Biol Chem. 1993 Feb 15;268(5):3389–3395. [PubMed] [Google Scholar]
- Middleton T., Sugden B. EBNA1 can link the enhancer element to the initiator element of the Epstein-Barr virus plasmid origin of DNA replication. J Virol. 1992 Jan;66(1):489–495. doi: 10.1128/jvi.66.1.489-495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins D. R., Milman G., Hayward S. D., Hayward G. S. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell. 1985 Oct;42(3):859–868. doi: 10.1016/0092-8674(85)90282-x. [DOI] [PubMed] [Google Scholar]
- Reisman D., Sugden B. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol Cell Biol. 1986 Nov;6(11):3838–3846. doi: 10.1128/mcb.6.11.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisman D., Yates J., Sugden B. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol Cell Biol. 1985 Aug;5(8):1822–1832. doi: 10.1128/mcb.5.8.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnos M., Zahn K., Inman R. B., Blattner F. R. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988 Feb 12;52(3):385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- Starr D. B., Hawley D. K. TFIID binds in the minor groove of the TATA box. Cell. 1991 Dec 20;67(6):1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- Su W., Middleton T., Sugden B., Echols H. DNA looping between the origin of replication of Epstein-Barr virus and its enhancer site: stabilization of an origin complex with Epstein-Barr nuclear antigen 1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10870–10874. doi: 10.1073/pnas.88.23.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden B., Marsh K., Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985 Feb;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Williams D. L., Kowalski D. Easily unwound DNA sequences and hairpin structures in the Epstein-Barr virus origin of plasmid replication. J Virol. 1993 May;67(5):2707–2715. doi: 10.1128/jvi.67.5.2707-2715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysokenski D. A., Yates J. L. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J Virol. 1989 Jun;63(6):2657–2666. doi: 10.1128/jvi.63.6.2657-2666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Guan N. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J Virol. 1991 Jan;65(1):483–488. doi: 10.1128/jvi.65.1.483-488.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J. L., Warren N., Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. 1985 Feb 28-Mar 6Nature. 313(6005):812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- Yates J., Warren N., Reisman D., Sugden B. A cis-acting element from the Epstein-Barr viral genome that permits stable replication of recombinant plasmids in latently infected cells. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3806–3810. doi: 10.1073/pnas.81.12.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]