Abstract
Recent studies have suggested that ubiquitination of mitochondrial proteins participates in regulating mitochondrial dynamics in mammalian cells, but it is unclear whether deubiquitination is involved in this process. Here, we identify human ubiquitin-specific protease 30 (USP30) as a deubiquitinating enzyme that is embedded in the mitochondrial outer membrane. Depletion of USP30 expression by RNA interference induced elongated and interconnected mitochondria, depending on the activities of the mitochondrial fusion factors mitofusins, without changing the expression levels of the key regulators for mitochondrial dynamics. Mitochondria were rescued from this abnormal phenotype by ectopic expression of USP30 in a manner dependent on its enzymatic activity. Our findings reveal that USP30 participates in the maintenance of mitochondrial morphology, a finding that provides new insight into the cellular function of deubiquitination.
INTRODUCTION
Mitochondria frequently fuse and divide to change their number, shape, and distribution within eukaryotic cells. These dynamic processes are important for a variety of biological functions such as cellular differentiation, inheritance of mitochondrial DNA (mtDNA), apoptosis, energy transmission, and meiosis and sporulation of yeast cells (Hales and Fuller, 1997; Nunnari et al., 1997; Frank et al., 2001; Skulachev, 2001; Gorsich and Shaw, 2004). After the initial discovery of the molecule responsible for mitochondrial fusion, called Fzo (fuzzy onions), in a Drosophila mutant defective in sperm development (Hales and Fuller, 1997), the key components of the mitochondrial fusion and division machinery were identified and characterized extensively in yeast and mammals. Mitochondrial fusion is mediated by the mitochondrial outer-membrane (MOM) GTPases mitofusin (Mfn)1 and Mfn2 (Fzo1p in yeast; Hermann et al., 1998; Rapaport et al., 1998; Santel and Fuller, 2001) and the intermembrane space-localized GTPase OPA1 (Mgm1p in yeast; Shepard and Yaffe, 1999; Wong et al., 2000; Olichon et al., 2003). Loss of any of their fusion activities induces mitochondrial fragmentation due to reduced rates of mitochondrial fusion (Hermann et al., 1998; Rapaport et al., 1998; Sesaki and Jensen 1999; Shepard and Yaffe, 1999; Wong et al., 2000; Chen et al., 2003; Eura et al., 2003; Olichon et al., 2003; Santel et al., 2003; Griparic et al., 2004; Lee et al., 2004). Mitochondrial fission is mediated by the cytosolic GTPase dynamin-related protein (Drp)1 (Dnm1p in yeast; Otsuga et al., 1998; Bleazard et al., 1999; Smirnova et al., 2001) and the integral MOM protein Fis1 (Fis1p in yeast; Mozdy et al., 2000; James et al., 2003; Yoon et al., 2003). Inhibition of either of their functions causes the formation of interconnected and elongated tubular mitochondria (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen 1999; Mozdy et al., 2000; Frank et al., 2001; Smirnova et al., 2001; Yoon et al., 2003; Lee et al., 2004; Stojanovski et al., 2004). Based on these findings, it has been considered that mitochondrial dynamics is tightly controlled by a balance of the activities of the mitochondrial fusion and division machinery. An increasing number of studies have identified additional factors influencing mitochondrial morphology and uncovered novel regulatory mechanisms for mitochondrial dynamics (for recent reviews, see Cerveny et al., 2007; Detmer and Chan, 2007; Hoppins et al., 2007). Recent studies have pointed out the involvement of ubiquitination in mitochondrial dynamics. In yeast, deficiencies of Rsp5p ubiquitin (Ub) ligase and subunits of the 26S proteasome cause abnormal mitochondrial morphology (Rinaldi et al., 1998, 2004; Fisk and Yaffe, 1999; Altmann and Westermann, 2005) and acceleration of proteasomal degradation of the fusion factor Fzo1p leads to mitochondrial fragmentation during the mating response (Neutzner and Youle, 2005; Escobar-Henriques et al., 2006). In mammals, the mitochondrial Ub ligase MARCH-V (also known as MITOL) seems to modulate mitochondrial division through ubiquitination of the division factors Drp1 and Fis1 (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007). However, the precise mechanism for the Ub-dependent regulation of mitochondrial dynamics remains elusive. Because ubiquitination can be rendered reversible by deubiquitinating enzymes (DUBs), a group of proteases that catalyze the cleavage of the isopeptide bond between Ub and its target protein, it is conceivable that the deubiquitination system would be used for the maintenance of mitochondrial morphology.
More than 90 DUBs are encoded in the human genome and 17 in the yeast genome, and they are classified into two major groups based on the sequence similarities of their DUB catalytic domains, that is, the Ub-specific protease (USP in human and UBP in yeast) and Ub C-terminal hydrolase (UCH) families. More than half of human DUBs and 16 yeast DUBs belong to the USP/UBP family, bearing little sequence similarity to each other beyond two short motifs, called the Cys and His boxes, which contain critical Cys and His residues for their DUB activities (Amerik and Hochstrasser, 2004; Nijman et al., 2005b). USPs/UBPs contain various extensions at their N and C termini and insertions in their catalytic domains; and these structural diversities are expected to contribute to substrate specificity and recognition, subcellular localization, and protein–protein interactions (Nijman et al., 2005b). An increasing number of studies have shown that USPs/UBPs play important roles in diverse cellular and physiological functions, including DNA repair (Nijman et al., 2005a), endocytic trafficking (Amerik et al., 2000; Mizuno et al., 2005), signal transduction (Brummelkamp et al., 2003; Kovalenko et al., 2003; Trompouki et al., 2003), transcriptional silencing (Moazed and Johnson, 1996; Henry et al., 2003), and cell growth and proliferation (Papa and Hochstrasser, 1993; Naviglio et al., 1998); but the specific role of DUBs in mitochondria has not been elucidated. In yeast, the MOM possesses the DUB Ubp16p, but neither deletion nor overexpression of the UBP16 gene results in obvious phenotypic defects in mitochondrial inheritance or morphology (Kinner and Kölling, 2003). However, it remains unclear as to whether a mitochondrial DUB exists in other organisms, especially mammals. In this study, we identified human USP30, a member of the USP family, as a novel mitochondrial DUB. The role of its DUB activity in mitochondria was addressed by RNA interference (RNAi) experiments. Our findings indicate that USP30 plays a role in regulating mitochondrial morphology.
MATERIALS AND METHODS
Plasmids
A cDNA encoding human USP30 was obtained from HeLa cells by reverse transcription-polymerase chain reaction amplification, and then it was inserted into the EcoRI–EcoRV sites of p3 × FLAGCMV-14 (Sigma-Aldrich, St. Louis, MO) to generate USP30-FLAG. FLAG-USP30ΔTM and glutathione transferase (GST)-USP30DUB were constructed by cloning the cDNA fragment encoding residues 57-517 of USP30 into the EcoRV site of p3 × FLAGCMV-10 (Sigma-Aldrich) and into the SmaI site of pGex4T1 (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), respectively. For generation of h/rUSP30, a partial cDNA fragment of rat USP30 was amplified from rat brain by RT-PCR with the primers 5′-tggcagatctcgtcctttgaggagcaggat-3′ and 5′-atctcctctgacctgtactctcgcccctgc-3′, and then it was inserted into the BglII–EcoRV sites of the USP30-FLAG plasmid. Myc-Fis1 was constructed by cloning the cDNA encoding mouse Fis1 into the EcoRI–XhoI sites of pcDNA3 (Invitrogen, Carlsbad, CA) encoding a 6 × Myc epitope tag. Matrix-targeted red fluorescent protein (mtRed; Nakamura et al., 2006) and Mfn2ΔTM (Honda et al., 2005) were described previously. All point mutations were introduced by primer-mediated site-directed mutagenesis. For generation of vectors expressing short-hairpin RNA (shRNA) targeted to human USP30, Drp1, Mfn1, and Mfn2, the following pairs of oligonucleotides with hairpin, terminator, and overhanging sequences were annealed and then inserted into the BamHI–HindIII sites of pSilencer2.1-U6 Puro vector (Ambion, Austin, TX): 5′-gatccgccagagtcctgttcgatttctcaagagaaaatcgaacaggactctggttttttggaaa-3′ and 5′-agcttttccaaaaaaccagagtcctgttcgattttctcttgagaaatcgaacaggactctggcg-3′ for shRNA#1; 5′-gatccgtcattacctcgtcattggctcaagagaccaatgacgaggtaatgacttttttggaaa-3′ and 5′-agcttttccaaaaaagtcattacctcgtcattggtctcttgagccaatgacgaggtaatgacg-3′ for shRNA#2; 5′-gatccgcagaagaatggggtaaatctcaagagaatttaccccattcttctgcttttttggaaa-3′ and 5′-agcttttccaaaaaagcagaagaatggggtaaattctcttgagatttaccccattcttctgcg-3′ for Drp1 RNAi; 5′-gatccgcgaaaccagatgaacctttctcaagagaaaaggttcatctggtttcgttttttggaaa-3′ and 5′-agcttttccaaaaaacgaaaccagatgaaccttttctcttgagaaaggttcatctggtttcgcg-3′ for Mfn1 RNAi; and 5′-gatccgcagtagtcctcaaggtttactcaagagataaaccttgaggactactgttttttggaaa-3′ and 5′-agcttttccaaaaaacagtagtcctcaaggtttatctcttgagtaaaccttgaggactactgcg-3′ for Mfn2 RNAi. The target sequences for Drp1 (Parone et al., 2006) and Mfn1 (Eura et al., 2003) RNAi were reported previously. The efficiency and specificity of shRNA-Mfn1 and shRNA-Mfn2 were demonstrated in Supplemental Figure S1. For control RNAi, pSilencer 2.1-U6 Puro Negative Control vector (Ambion) was used. The cDNA sequences of all constructs were verified by sequencing.
Cell Culture, Fluorescence Microscopy, and Quantification of Mitochondrial Morphology
Maintenance of HeLa and COS7 cells, transient and stable transfection with plasmids, and immunofluorescence staining were performed as described previously (Nakamura et al., 2005). The expression plasmids (i.e., shRNA-Drp1, Myc-Fis1, Mfn2ΔTM, h/rUSP30, and h/rUSP30-CS) were cotransfected at a 10:1 ratio with the mtRed plasmid. The shRNA-Mfn1 and shRNA-Mfn2 plasmids were cotransfected at a 10:10:1 ratio with the mtRed plasmid. Images were captured with an inverted fluorescence microscope (IX70; Olympus, Tokyo, Japan) equipped with a 100× oil immersion objective and a SenSys charge-coupled device camera (Photometrics, Huntington Beach, CA), and then they were analyzed with MetaVue software (Molecular Devices, Sunnyvale, CA). Transiently transfected cells were used for experiments at 24 h after transfection, with the exception of Drp1 RNAi (48 h) and Mfn1 and Mfn2 RNAi (72 h). Quantitative analyses of mitochondrial morphology were performed by counting 100–200 randomly selected cells for each experiment. Cells were scored as having elongated mitochondria when >50% of mitochondria in the image were more than 8 μm in length; cells were scored as having fragmented mitochondria when >50% of mitochondria in the image were <1 μm in length. Data were presented as the mean ± SD of three independent experiments. Statistical analysis was carried out by using Student's t test. A value of p < 0.05 was considered significant.
Northern Blot Analysis
Total RNA was prepared from rat tissues with Isogen (Nippon Gene, Tokyo, Japan). Poly(A)+ RNA was purified from cultured cells with an illustra QuickPrep MicromRNA purification kit (GE Healthcare). After separation, the total RNA (20 μg) and mRNA (2 μg) were transferred to a Hybond-N+ nylon membrane (GE Healthcare). The filters were then hybridized with 32P-labeled cDNA probes in PerfectHyb hybridization buffer (Toyobo, Osaka, Japan) at 68°C overnight. The filters were subsequently washed with 1× SSC (0.15 M NaCl, 15 mM sodium citrate, pH 7.5, and 0.1% SDS) at 68°C, and then they were exposed to an imaging plate (FujiFilm, Tokyo, Japan). Signals were analyzed with a BAS2000 bio-imaging analyzer (FujiFilm).
Antibodies
Rabbit polyclonal antisera against Mfn1 (Nakamura et al., 2006), Mfn2 (Honda and Hirose, 2003), and MARCH-V (Nakamura et al., 2006) were described previously. Polyclonal antiserum specific for Fis1 was made in rabbits that had been immunized with the recombinant His6-tagged protein of the N-terminal portion of mouse Fis1 (residues 1-120). Other antibodies against the following antigens were also used: FLAG (clone M2) and α-tubulin (clone B-5-1-2; Sigma-Aldrich); Ub (clone Ubi-1; Zymed Laboratories, South San Francisco, CA); calreticulin (Affinity BioReagents, Golden, CO); and syntaxin 6 (clone 30), Drp1 (clone 8), OPA1 (clone 18), Tom20 (clone 29), and Tim23 (clone 32; BD Transduction Laboratories, Lexington, KY).
Membrane Extraction Assay
COS7 cells transfected with USP30-FLAG were homogenized in 500 μl of phosphate-buffered saline (PBS) containing protease inhibitors (10 mM leupeptin, 1 mM pepstatin, 5 mg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride). The homogenate was centrifuged at 60,000 rpm in a TLA100.3 rotor (Beckman Coulter, Fullerton, CA) for 20 min. The supernatant (the cytosolic fraction) was recovered, and the membrane pellet was resuspended and homogenized in 500 μl of PBS containing protease inhibitors. Then, the membrane homogenates were divided into four aliquots of 100 μl each, and they were centrifuged at 60,000 rpm for 20 min. The resulting membrane pellets were incubated in either PBS, 1 M NaCl, 0.1 M Na2CO3, pH 11.5, or 1% Triton X-100 in PBS (150 μl each) on ice for 1 h. After the samples had been centrifuged at 60,000 rpm for 20 min; the supernatants were recovered as soluble fractions (fraction S); and the pellets were washed by resuspending them in PBS, and then they were centrifuged at 60,000 rpm for 20 min. The resulting pellets were then extracted in 150 μl of radioimmunoprecipitation assay buffer (10 mM sodium phosphate, pH 7.4, containing 0.15 M NaCl, 1% Nonidet P-40, 5% sodium deoxycholate, 0.1% SDS, 2 mM EDTA, 50 mM NaF, and protease inhibitors). These extracts were centrifuged for 20 min at 60,000 rpm, and the resulting supernatant was recovered as the extracted pellet fraction (fraction P).
Subcellular Fractionation and Proteinase K Digestion
COS7 cells transfected with USP30-FLAG were homogenized in 6 ml of SHP buffer (10 mM HEPES-KOH, pH 7.0, containing 0.25 M sucrose, and 0.2 M KCl) supplemented with protease inhibitors as used for the membrane extraction assay, and then they were centrifuged at 3000 × g for 10 min. The supernatant was centrifuged at 6800 rpm in an SW41Ti rotor (Beckman Coulter) for 12 min, yielding the mitochondria-rich pellet and the postmitochondrial supernatant. The mitochondria-rich pellet was resuspended in 5 ml of SHP buffer, and then it was centrifuged at 6800 rpm in an SW41Ti rotor for 12 min. The resulting pellet was used as the mitochondrial-rich fraction. The postmitochondrial supernatant was centrifuged at 24,000 rpm in an SW41Ti rotor for 20 min, yielding the cytosolic supernatant and the postmitochondrial membrane pellet. The postmitochondrial membrane pellet was washed by resuspension in SHP buffer followed by centrifugation at 24,000 rpm in an SW41Ti rotor for 20 min. The resulting pellets and the mitochondrial membrane fraction were then extracted in 100 μl of 1% Triton X-100 in PBS, and they were used for Western blot analysis. For a proteinase K digestion assay, the mitochondria-rich fraction was rinsed with SHP buffer, and then it was incubated on ice for 1 h in the presence or absence of 0.1 mg/ml proteinase K in SHP buffer. After the protease reaction had been quenched by adding phenylmethylsulfonyl fluoride (final concentration 1 mM), the samples were centrifuged for 30 min at 24,000 rpm in an SW41Ti rotor. The pellets were extracted with Laemmli sample buffer.
In Vitro DUB Assay
Lys-48– or Lys-63–linked tetra-Ub chains (Ub4) chains (50 ng; BostonBiochem, Cambridge, MA) were incubated at 37°C for 24 h in 50 μl of PBS, pH 7.4, containing 5 mM MgCl2 and 2 mM dithiothreitol in the presence or absence of 20 μg of GST-USP30DUB or GST-USP30DUB-CS. The reaction was quenched by adding 12 μl of 5× concentrated Laemmli buffer (10% SDS, 0.3 M Tris-HCl, pH 6.8, 40% glycerol, 5% 2-mercaptoethanol, and 0.025% bromphenol blue). The samples (20 μl) were separated on a 18% SDS-polyacrylamide gel, and then they were transferred to a polyvinylidene difluoride membrane (Immobilon-PSQ [0.2-μm pore size]; Millipore, Billerica, MA) by using a tank-blotting apparatus (Atto, Tokyo, Japan) in 25 mM Tris, 192 mM glycine, and 20% methanol at 14 V for 5 h at 10°C. Ub molecules were detected by incubation with anti-Ub antibody (1 μg/ml in Tris-buffered saline/Tween 20 [0.05% Tween 20, 150 mM NaCl, and 10 mM Tris-HCl, pH 7.6]) followed by enhanced chemiluminescence (ECL Plus; GE Healthcare) with exposure to Hyperfilm-ECL (GE Healthcare) for 1 h.
RESULTS
USP30 Is a Novel Transmembrane DUB of the MOM
By hydropathy plot analysis of currently available DUB protein sequences from the GenBank database, we found that USP30 had a membrane topology, albeit with weak protein sequence homology, similar to that of yeast Ubp16p—an N-terminally anchored protein with a cytosolic DUB catalytic domain (Figure 1A; Kinner and Kölling, 2003). Human USP30 consisted of 517-amino acid residues with a hydrophobic region at its N terminus (residues 35-54) followed by a DUB catalytic domain (residues 69-499) containing conserved Cys and His residues critical for the DUB activity (Figure 1B; GenBank accession no. AJ586136). BLAST searching revealed the presence of its USP30 homologues with similar structural properties in other mammals, birds, amphibians, fish, insects, and worms. Based on their structural and topological similarities, we predicted that USP30 would serve as the mammalian orthologue of Ubp16p, and put this to the test with a phylogenetic analysis by using all UBP proteins in yeast. The result of this analysis showed that the USP30 members were closely related to Ubp16p in the yeast UBP family (Figure 1C). Northern blot analysis revealed broad tissue expression of a 3-kb rat USP30 mRNA with relatively higher yields in brain, testis, and ovary; and an additional smaller transcript (2.5 kb) was detected in testis (Figure 1D), suggesting a generalized function in many cell types and a more specific role in brain and reproductive organs.
Figure 1.
Structural features, phylogenetic relationship, and tissue distribution of USP30. (A) A Kyte–Doolittle hydropathy plot of the amino acid sequence of human USP30. The plot was constructed by using the GENETYX program (Software Development, Tokyo, Japan). The putative TM region is shaded. (B) Schematic structure of human USP30. The putative TM region and DUB catalytic domain are indicated in black and gray, respectively. The conserved Cys (Cys 77) and His (His 452) residues are indicated by C and H, respectively. (C) A phylogenetic analysis of human USP30 with its homologues and the yeast (S. cerevisiae) UBP members. The tree was constructed by the neighbor-joining method by using the ClustalW computer program. The scale bar represents a genetic distance of 0.5-amino acid substitution per site. The GenBank accession numbers are as follow: human USP30, CAE51936; rat (Rattus norvegicus) USP30, NP_001100623; chicken (Gallus gallus) USP30, XP_415187; western clawed frog (Xenopus tropicalis) USP30, AAI35926; zebrafish (Danio rerio) USP30, CAM16313; fruit fly (Drosophila melanogaster) USP30 #1 (CG3016-PA), NP_572274; fruit fly USP30 #2 (GA15687-PA), XP_001355317; nematode (Caenorhabditis elegans) USP30 (Y67D2.2), NP_497422; UBP1p, NP_010161; UBP2p, NP_014767; UBP3p, NP_011078; UBP4p, NP_010354; UBP5p, NP_011071; UBP6p, NP_116665; UBP7p, NP_012110; UBP8p, NP_013950; UBP9p, NP_011024; UBP10p, NP_014213; UBP11p, NP_013024; UBP12p, NP_012338; UBP13p, NP_009486; UBP14p, NP_009614; UBP15p, NP_014033; and UBP16p, NP_015253. (D) A Northern blot of 20 μg of total RNA prepared from the indicated rat tissues was hybridized with specific probes for USP30 (top) and β-actin (bottom).
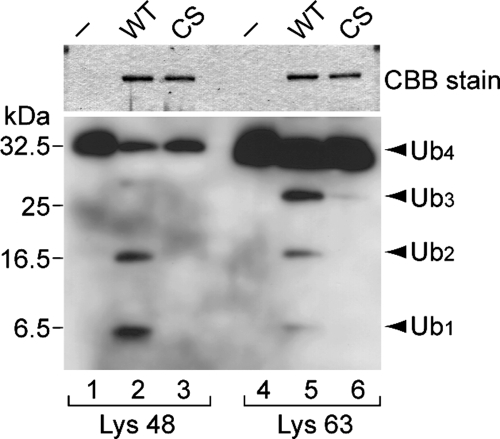
Quesada et al. have demonstrated the DUB activity of USP30 against the Ub–β-galactosidase fusion protein (Quesada et al., 2004). To confirm the USP30 activity, we incubated purified recombinant GST-fusion proteins of the USP30 DUB catalytic domain (residues 57-517; GST-USP30DUB) with Ub4 chains. Western blotting of the reaction samples with anti-Ub antibody revealed that GST-USP30DUB cleaved both Lys-48- and Lys-63-linked Ub4, yielding their trimer, dimer, and monomer (Figure 2, lanes 2 and 5). This DUB activity was abolished by a point mutation of a conserved cysteine (Cys 77) to Ser (GST-USP30DUB-CS; Figure 2, lanes 3 and 6), indicating that Cys 77 is essential for the DUB activity.
Figure 2.
DUB activity of USP30. In vitro deubiquitination assays of Lys-48– (lanes 1–3) and Lys-63–linked (lanes 4–6) Ub4 were performed in the absence (−; lanes 1 and 4) or presence of GST-USP30DUB (WT; lanes 2 and 5) or GST-USP30DUB-CS (CS; lanes 3 and 6). The reaction samples were analyzed by SDS-PAGE followed by Coomassie Brilliant Blue staining to detect the GST-fusion proteins used (top) and Western blotting with anti-Ub antibody (bottom). Arrowheads indicate the positions of tetra- (Ub4), tri- (Ub3), di- (Ub2), and mono-Ub (Ub1).
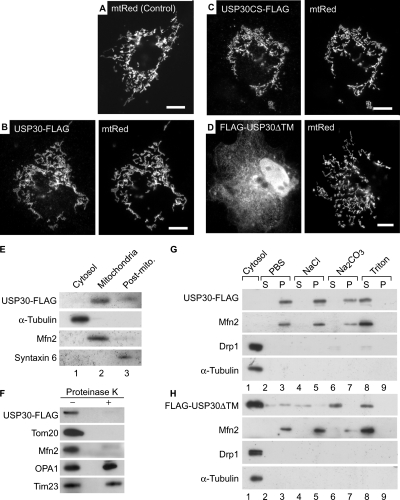
To examine whether USP30 was localized to mitochondria, COS7 cells were transfected with a C-terminally FLAG-tagged USP30 (USP30-FLAG) together with an mtRed to visualize mitochondria (Figure 3A). Immunofluorescence microscopy using anti-FLAG antibody demonstrated a clear mitochondrial localization of USP30-FLAG (Figure 3B). A mutant USP30 containing the C77S substitution (USP30CS-FLAG) was also detected on mitochondria (Figure 3C), indicating that the DUB activity is not required for the mitochondrial localization of USP30. Furthermore, a deletion of 56 amino-acid residues at its N terminus resulted in a cytoplasmic distribution of the protein (FLAG-USP30ΔTM; Figure 3D). The mitochondrial localization of USP30-FLAG was confirmed by subcellular fractionation of the transfected COS7 cells. The cytosolic, mitochondrial, and postmitochondrial fractions were subjected to Western blot analysis with antibodies against FLAG, α-tubulin (a cytosolic protein), Mfn2 (a mitochondrial protein), and syntaxin 6 (a Golgi and endosomal protein). As shown in Figure 3E, USP30-FLAG was detected predominantly in the mitochondria-rich fraction. To determine the submitochondrial localization of USP30, we treated intact mitochondria from COS7 cells expressing USP30-FLAG with proteinase K, which digests the cytosolic parts of MOM proteins. As shown in Figure 3F, the proteinase K treatment digested the MOM proteins (Tom20 and Mfn2), whereas the intermembrane-space and inner-membrane proteins (OPA1 and Tim23) were protected. Under these conditions, USP30-FLAG was not detected in the proteinase K-treated mitochondria with anti-FLAG antibody (Figure 3F, top), suggesting that USP30 was localized to the MOM with its C terminus protruding toward the cytosol. Subcellular fractionation experiments revealed that USP30-FLAG was present in the membrane fraction (Figure 3, E and G) and that it could be solubilized effectively in 1% Triton X-100 but not in PBS, high-salt solution, or alkaline carbonate. This extraction profile was similar to that of the integral membrane protein Mfn2 (Figure 3G). In contrast, the majority of FLAG-USP30ΔTM was fractionated into the cytosolic fraction (Figure 3H, top, lane 1), consistent with the observation by immunofluorescence microscopy (Figure 3D). Residual membrane-associated FLAG-USP30ΔTM was released mostly from membranes in high-salt solution and alkaline carbonate (Figure 3H). These results indicate that USP30 is an integral membrane protein and that its N-terminal hydrophobic region forms a transmembrane (TM) span. Together, the above-mentioned results demonstrate that USP30 is a DUB anchored to the MOM.
Figure 3.
Subcellular localization of USP30. (A–D) COS7 cells transiently transfected with matrix-targeted red fluorescent protein (mtRed) alone (Control; A) or together with USP30-FLAG (B), USP30CS-FLAG (C) or FLAG-USP30ΔTM (D) were stained with anti-FLAG antibody. Signals for anti-FLAG antibody (left) and mtRed (right) are shown. Bars, 10 μm. (E) The cytosolic, mitochondrial, and postmitochondrial (Post-mito.) fractions (5 μg of protein each) were analyzed by Western blotting with antibodies against FLAG, α-tubulin, Mfn2, and syntaxin 6. (F) The mitochondria-rich fractions of COS7 cells expressing USP30-FLAG were incubated with (+) or without (−) 100 μg/ml proteinase K and then used for Western blotting with antibodies against the indicated antigens. (G and H) Homogenates of COS7 cells expressing USP30-FLAG (G) or FLAG-USP30ΔTM (H) were fractionated into the cytosolic (lane 1) and membranous fractions. The membranes were incubated with PBS (lanes 2 and 3), 1 M NaCl (lanes 4 and 5), 0.1 M Na2CO3 (pH 11.5, lanes 6 and 7), or 1% Triton X-100 (lanes 8 and 9) followed by ultracentrifugation to separate soluble protein supernatants (S; lanes 2, 4, 6, and 8) from membranous pellets (P; lanes 3, 5, 7, and 9). The samples were analyzed by Western blotting with antibodies against FLAG, Mfn2 (a TM protein), Drp1 (a cytosolic protein), and α-tubulin (a cytosolic protein).
Mitochondrial Targeting Sequences of USP30
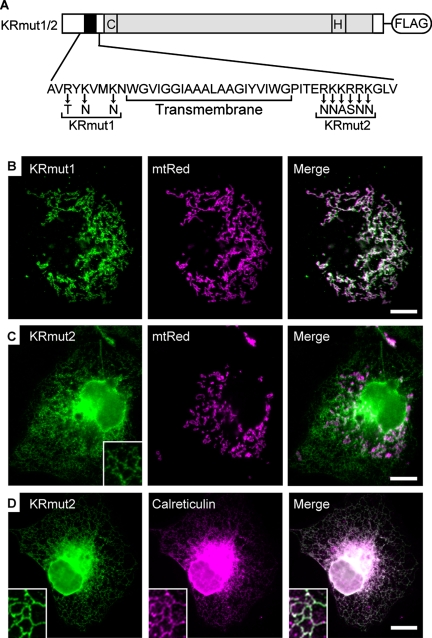
The mitochondrial targeting and membrane insertion of several N-terminally anchored MOM proteins, such as Tom20 and Tom70, require their TM span and positively charged residues in its flanking regions, called “signal-anchor” sequences (Kanaji et al., 2000; Suzuki et al., 2002; Rapaport 2003). USP30 also possessed positively charged residues in both N- and C-terminal regions flanking the TM span (Figure 4A). To examine whether these sequences were important for mitochondrial targeting of USP30, we replaced the positively charged residues in each sequence with uncharged residues (Figure 4A). Charge neutralization in the region before the TM span did not affect the mitochondrial localization of USP30 (KRmut1; Figure 4B). In contrast, a mutant USP30 with replacements in the region after the TM span (KRmut2) was not localized to the mitochondria (Figure 4C), but rather it was redistributed into a perinuclear and cytoplasmic reticular pattern that colocalized with the endoplasmic reticulum marker calreticulin (Figure 4D). Thus, these results indicate that mitochondrial targeting of USP30 requires a net positive charge in the C-terminal TM flanking region as well as the TM span.
Figure 4.
Mitochondrial targeting of USP30. (A) Schematic structure of KRmut1 and 2. The amino acid sequence of and around the putative TM region (residues 26-67) and those sequences of KRmut1 and 2 mutants are shown. (B and C) COS7 cells transiently transfected with mtRed together with KRmut1 (B) or KRmut2 (C) were stained with anti-FLAG antibody. Signals for anti-FLAG antibody (left) and mtRed (middle) are shown. Inset in C shows a higher magnification image in the peripheral region of the cell. Bars, 10 μm. (D) COS7 cells transiently transfected with KRmut2 were double stained with antibodies against FLAG and calreticulin. Signals for anti-FLAG antibody (left) and anti-calreticulin antibody (middle) are shown. Insets show a higher magnification image in the peripheral region of the cell. Bar, 10 μm.
USP30 Knockdown Leads to Mitochondrial Elongation
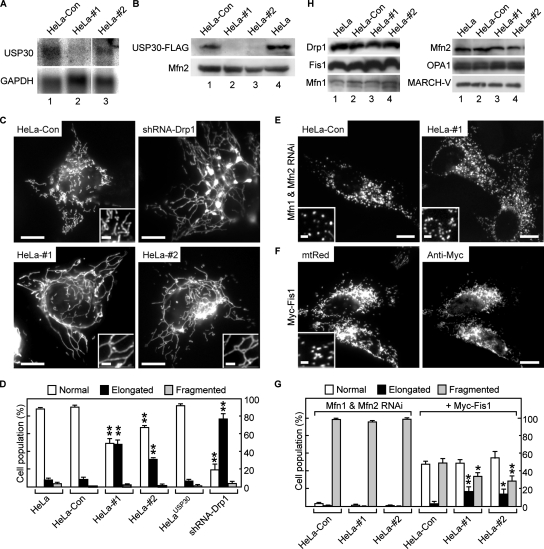
Next, we investigated whether USP30 had any impact on mitochondrial morphology by using RNAi to deplete USP30. HeLa cells were stably transfected with vectors expressing shRNAs against human USP30 (shRNA#1 and #2) or with a control nonsilencing shRNA (termed HeLa-#1, HeLa-#2, and HeLa-Con cells, respectively). The levels of USP30 mRNA were reduced in HeLa-#1 and HeLa-#2 to ∼28 and 60%, respectively, of those in HeLa-Con cells (Figure 5A; band intensities were quantified with ImageJ software and normalized to glyceraldehyde-3-phosphate dehydrogenase [GAPDH] levels). Additionally, the expression of exogenous USP30-FLAG proteins was also suppressed by the USP30 shRNAs (Figure 5B). In 30–50% of the USP30-depleted cells, their mitochondria exhibited an aberrant morphology, being much longer and more highly branched than mitochondria in the HeLa-Con cells (Figure 5, C and D). This phenotype, however, was less severe than that in HeLa cells after shRNA-mediated depletion of Drp1, a mitochondrial division factor (Figure 5, C, top right, and D; 77.0 ± 6.1% of Drp1-depleted cells showed mitochondrial elongation). Because mitochondrial morphology is maintained by balanced fusion and fission events, we addressed whether the effect of USP30 RNAi would be affected by a blockage of fusion or an enhancement of fission. Depletion of two mitochondrial fusion factors, Mfn1 and Mfn2, causes an extensive fragmentation of mitochondria (Chen et al., 2003; Eura et al., 2003). The expression vectors of shRNA against Mfn1 and Mfn2, together with the mtRed vector, were used to transfect HeLa-Con and USP30-depleted cells. As expected, most of the transfected HeLa-Con cells exhibited mitochondrial fragmentation 72 h after transfection (Figure 5E, left, and G). Under these conditions, in the transfected USP30-depleted cells, the mitochondrial elongation induced by USP30 RNAi was clearly blocked; and mitochondrial fragmentation was observed at rates similar to those for HeLa-Con cells (Figure 5E, right, and G; 98.7 ± 1.2% of HeLa-#1 cells and 99.0 ± 0.9% of HeLa-#2 cells vs. 95.9 ± 0.9% of HeLa-Con cells). In addition, similar results were obtained by overexpressing Mfn2ΔTM, the TM-less Mfn2 mutant that blocks mitochondrial fusion (Honda et al., 2005; Supplemental Figure S2). Thus, these results suggest that mitochondrial elongation induced by USP30 RNAi depends on fusion events mediated by Mfn proteins. Next, we examined the effects of overexpression of the mitochondrial division factor Fis1, which also causes fragmentation of mitochondria (Yoon et al., 2003). Exogenous expression of Myc-tagged Fis1 (Myc-Fis1) resulted in mitochondrial fragmentation in USP30-depleted cells (Figure 5F), but the cell populations with this phenotype were lower in number than that population of HeLa-Con cells expressing Myc-Fis1 (Figure 5G, gray bar; 34.0 ± 3.7% of HeLa-#1 cells and 28.7 ± 5.7% of HeLa-#2 cells vs. 49.2 ± 4.5% in HeLa-Con cells); and the elongated mitochondria were still observed in USP30-depleted cells (Figure 5G, filled bar; 16.9 ± 4.8% of HeLa-#1 cells and 14.0 ± 5.5% of HeLa-#2 cells vs. 3.0 ± 1.8% of HeLa-Con cells). These results suggest that an increase in the fission activities counteracts the effect of USP30 knockdown on mitochondrial morphology. We next examined whether overexpression of USP30 would have any effect on mitochondrial morphology. In contrast to the results of USP30 knockdown, the overexpression of USP30-FLAG did not lead to any apparent change in mitochondrial shape, as seen in transiently transfected COS7 cells (Figure 3B) or in stably transfected HeLa cells (HeLaUSP30 cells; Figure 5D).
Figure 5.
Effect of USP30 knockdown on mitochondrial morphology. (A) Northern blot of 2 μg of poly(A)+ RNA purified from HeLa-Con (lane 1), HeLa-#1 (lane 2) or HeLa-#2 (lane 3) cells was hybridized with specific probes for USP30 (top) and GAPDH (bottom). (B) HeLa-Con (lane 1), HeLa-#1 (lane 2), HeLa-#2 (lane 3), and parental HeLa (lane 4) cells were transiently transfected with USP30-FLAG. The membrane fractions of the cells (20 μg of protein) were analyzed by Western blotting with antibodies against FLAG (top) and Mfn2 (bottom). (C) The mtRed-expression vector was used to transfect HeLa-Con (top left), HeLa-#1 (bottom left), and HeLa-#2 (bottom right) cells or to cotransfect HeLa cells with shRNA-Drp1 (top right). The cells were analyzed by fluorescence microscopy 24 h (HeLa-Con, HeLa-#1, and HeLa-#2 cells) or 48 h (shRNA-Drp1) after transfection. Signals for mtRed are shown. Insets show higher magnification images in the peripheral regions of the cells. Bars, 10 and 2 μm in the insets. (D) The bar graph shows the percentage of HeLa, HeLa-Con, HeLa-#1, HeLa-#2, HeLaUSP30, and HeLa cells expressing shRNA-Drp1 having the indicated mitochondrial morphology. Data are expressed as the means ± SD of triplicate. **p < 0.01 compared with the same morphology in HeLa-Con cells. (E) HeLa-Con (left) and HeLa-#1 (right) cells were transiently transfected with mtRed together with Mfn1 and Mfn2 shRNAs. The cells were analyzed by immunofluorescence microscopy 72 h after transfection. Signals for mtRed are shown. Insets show higher magnification images in the peripheral regions of the cells. Bars, 10 and 2 μm in the insets. (F) HeLa-#1 cells were transiently transfected with mtRed together with Myc-Fis1. At 24 h after transfection, the cells were stained with anti-Myc antibody and then observed by immunofluorescence microscopy. Signals for mtRed (left) and anti-Myc antibody (right) are shown. The inset shows a higher magnification image in the peripheral region of the cell. Bars, 10 and 2 μm in the inset. (G) The bar graph shows the percentage of the designated cells expressing Mfn1 and Mfn2 shRNAs (Mfn1 and Mfn2 RNAi) or Myc-Fis1 and having the indicated mitochondrial morphology. Data are expressed as the means ± SD of triplicate samples. *p < 0.05 and **p < 0.01 compared with the same morphology in HeLa-Con cells under the same conditions. (H) The membrane fractions (20 μg of protein) of HeLa (lane 1), HeLa-Con (lane 2), HeLa-#1 (lane 3), and HeLa-#2 (lane 4) cells were analyzed by Western blotting by using antibodies specific for the indicated antigens.
Given the earlier evidence that the Ub-proteasomal system modifies mitochondrial morphology through regulating the stability of Drp1, Fis1, and Fzo1p (Neutzner and Youle, 2005; Escobar-Henriques et al., 2006; Yonashiro et al., 2006), we assessed whether USP30 knockdown would lead to a change in the protein levels of the central mediators of mitochondrial dynamics (i.e., Drp1, Fis1, Mfn1, Mfn2, and OPA1). Western blotting of the membrane fractions of HeLa-#1 and HeLa-#2 cells showed that their protein levels were not apparently altered compared with those in parental HeLa and HeLa-Con cells (Figure 5H), thus suggesting that USP30 did not regulate the protein turnover of these regulatory proteins. Likewise, we did not detect any significant change in the steady-state protein levels of MARCH-V (Figure 5H, bottom right), which has been shown to undergo proteasomal degradation through autoubiquitination (Yonashiro et al., 2006). Together, these results indicate that USP30 expression is required for the maintenance of balanced mitochondrial morphology.
DUB Activity of USP30 Is Required for Normal Mitochondrial Morphology
To confirm the specificity of the RNAi effect, we performed a rescue experiment with a chimeric construct of human USP30 in which the region of residues 152-517 was replaced with the corresponding region of rat USP30 (h/rUSP30; Figure 6A). The nucleotide sequence of rat USP30 differs from human USP30 at several positions in the RNAi target sites (Figure 6B). When USP30-deficient cell lines were transfected with h/rUSP30, its expression was somewhat suppressed but displayed resistance to USP30 RNAi (Figure 6C). Under these conditions, the aberrant mitochondrial morphology was restored to the control state (Figure 6D, top, and E), suggesting that h/rUSP30 complements the function of USP30. Next, to examine whether this recovery was mediated by the DUB activity of USP30, we introduced the C77S point mutation into h/rUSP30 to inactivate its DUB activity (h/rUSP30-CS). On ectopic expression of h/rUSP30-CS, the cell population with mitochondrial elongation was slightly, although not significantly, reduced but still remained at a level similar to that of nontransfected USP30-depleted cells (Figure 6D, bottom, and E; compare with Figure 5D; 39.5 ± 3.9% of HeLa-#1 cells expressing h/rUSP30-CS vs. 48.3 ± 5.6% of HeLa-#1 cells, p = 0.090; 26.6 ± 7.1% of HeLa-#2 cells expressing h/rUSP30-CS vs. 31.3 ± 0.9% of HeLa-#2 cells, p = 0.32). These results indicate that the DUB activity of USP30 is critical for its impact on mitochondrial morphology.
Figure 6.
Rescue of USP30 knockdown phenotype with an RNAi-resistant enzymatically active USP30. (A) A schematic structure of h/rUSP30. The C-terminal portion of USP30-FLAG (residues 152-517) was replaced with the rat USP30 sequence. Arrows indicate the regions corresponding to the USP30 RNAi sites of human USP30 mRNA (#1, shRNA#1; #2, shRNA#2). (B) Nucleotide sequence alignments of the shRNA#1 and shRNA#2 target sequences of human USP30 and their corresponding regions of rat USP30. (C) HeLa-Con (lanes 1 and 4), HeLa-#1 (lanes 2 and 5), and HeLa-#2 (lanes 3 and 6) cells were transiently transfected with h/rUSP30 (lanes 1–3) or h/rUSP30-CS (lanes 4–6). The membrane fractions of the cells (20 μg of protein) were analyzed by Western blotting with antibodies against FLAG (top) and Mfn2 (bottom). (D) HeLa-#2 cells were transiently transfected with mtRed together with h/rUSP30 (top) or h/rUSP30-CS (bottom), and then they were examined by immunofluorescence microscopy with anti-FLAG antibody. Signals for mtRed (left) and anti-FLAG antibody (right) are shown. Insets show higher magnification images in the peripheral regions of the cells. Bars, 10 μm. (E) The bar graph shows the percentage of the designated cells, expressing the indicated chimeric proteins that showed the various mitochondrial morphologies. Data are expressed as the means ± SD of triplicate samples. *p < 0.05 and **p < 0.01 compared with the same morphology in HeLa-Con cells under the same conditions.
DISCUSSION
Earlier genetic studies in yeast have suggested that ubiquitination is involved in maintaining the structural and functional integrity of mitochondria (Rinaldi et al., 1998, 2004; Fisk and Yaffe, 1999; Altmann and Westermann, 2005; Neutzner and Youle, 2005; Escobar-Henriques et al., 2006). Recent studies in mammalian cells have revealed that the mitochondrial Ub ligase MARCH-V seems to regulate the mitochondrial dynamics by ubiquitinating Drp1 and Fis1 (Nakamura et al., 2006; Yonashiro et al., 2006; Karbowski et al., 2007). However, little is known as to how ubiquitination of mitochondrial proteins is regulated. Hence, this study was initiated with the objective to identify a mammalian DUB protein responsible for this regulation. Database mining in combination with hydropathy plot analysis identified human USP30 as a TM-containing DUB member topologically similar to the yeast mitochondrial DUB Ubp16p. Expectedly, USP30 was found to be anchored to the MOM by its N-terminal TM domain with its C-terminal DUB catalytic domain facing the cytosol (Figure 3). The membrane orientation of USP30 suggests that it could catalyze deubiquitination of components of the MOM and cytosolic proteins in vicinity to mitochondria. The deletion and mutagenesis analyses demonstrated that the TM domain and the positively charged residues in its C-terminal flanking region are required for the targeting and insertion of USP30 into the mitochondrial membrane (Figures 3 and 4), suggesting that these structures function as a signal-anchor sequence. Possibly, the MOM import of USP30 may be mediated via a mechanism similar to that for other N-terminal–anchored MOM proteins such as Tom20 and Tom70 (Suzuki et al., 2002; Ahting et al., 2005).
The most notable finding of this study is that depletion of USP30 expression led to elongated and interconnected mitochondria. Because this mitochondrial elongation was effectively attenuated by an enhancement of Fis1-mediated fission (Figure 5, F and G), the balance between mitochondrial fusion and fission was likely shifted toward fusion in the USP30-depleted cells, which shift may have been due to a decrease in fission and/or an increase in fusion. Indeed, the phenotype caused by USP30 RNAi closely resembles those phenotypes obtained by depleting Drp1 or Fis1 expression (Lee et al., 2004; Stojanovski et al., 2004). However, USP30 per se seems unlikely to act as the division machinery, because the mitochondrial defect in the USP30-depleted cells was considerably milder than that caused by Drp1 RNAi (Figure 5, C and D). The formation of elongated mitochondria was most likely dependent on the activity of the fusion machinery, because depletion of both Mfn1 and Mfn2 expression counteracted almost completely the effect of USP30 RNAi, and they induced mitochondrial fragmentation in most of the transfected cells (Figure 5, E and G). Based on these findings, we conclude that USP30 contributes to, but is not indispensable for, the maintenance of mitochondrial morphology by acting on the mitochondrial fusion and fission pathways. The rescue experiments of USP30 knockdown demonstrated that the DUB activity is essential for the action of USP30 (Figure 6). This finding allowed us to expect that overexpression of wild-type USP30 would lead to its overactivity with a diametrically opposite effect—mitochondrial fragmentation. However, the overexpression of USP30-FLAG did not have any discernible effect on mitochondrial morphology (Figures 3B and 5D), suggesting that endogenous levels of the USP30 activity were sufficient for maintaining mitochondrial morphology and that its higher expression was well tolerated. We also observed no significant change in mitochondrial morphology in cells overexpressing the C77S mutant USP30 (USP30CS-FLAG; Figure 3C) or the soluble USP30 form lacking the TM region (FLAG-USP30ΔTM; Figure 3D). These mutant proteins seem not to enhance or interfere with the endogenous USP30 function, and thus a certain DUB activity toward mitochondria would enable USP30 to influence the regulation of mitochondria dynamics.
Currently, we cannot clearly explain the mechanism by which depletion of USP30 activity caused mitochondrial elongation. It has become apparent that Ub modification is used for purposes other than a signal for proteasomal degradation, such as changing the activities and subcellular localization of substrate proteins, which is attributed at least to many variations in its conjugation patterns (e.g., poly-, mono-, and multiubiquitiation and Lys-48– and Lys-63–linked Ub chains; Pickart and Fushman, 2004). In some cases, DUBs reversely regulate the Ub-mediated nonproteasomal processes, including endosomal protein trafficking by deubiquitinating TM cargos (Amerik et al., 2000; Mizuno et al., 2005) and transcriptional regulation by deubiquitinating histone (Henry et al., 2003). Possibly, USP30 may control the activities of the regulators for mitochondrial dynamics in a similar way. In this regard, it is notable that MARCH-V Ub ligase seems to regulate Drp1-mediated mitochondrial division without affecting the stability of Drp1, Fis1 or Mfn2 (Karbowski et al., 2007). Of course, we cannot exclude the possibility that USP30 controls the stability and/or activity of yet unidentified Ub-substrates that influence mitochondria dynamics. The structural similarity and mitochondrial localization imply that USP30 may be a mammalian orthologue of yeast Ubp16p, but it is still unclear whether these two proteins are functionally related. One possibility is that USP30 and Ubp16p may have an unidentified conserved function and that USP30 would have acquired the ability to alter mitochondrial morphology during evolution.
In conclusion, we have demonstrated that USP30 is a DUB anchored to the MOM and that it contributes to the regulatory mechanism for mitochondrial dynamics in mammalian cells. Loss of its activity disturbs the maintenance of mitochondrial morphology and it leads to mitochondrial elongation. Our findings not only underscore the importance of Ub modification on mitochondrial dynamics but also uncover a new function of the DUB protein family. The identification and characterization of the relevant substrates of ubiquitination and deubiquitination should help us to clarify the exact function of USP30 and the molecular basis of the structural and functional integrity of mitochondria.
Supplementary Material
ACKWNOWLEDGMENTS
We thank Dr. Shinji Honda and Setsuko Sato for technical and secretary assistance and Dr. Larry Frye for copyediting the manuscript. This work was supported by Grant-in-Aid for Scientific Research 14104002 and by the 21st Century Centers of Excellence (COE) and Global COE Programs of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations used:
- Drp1
dynamin-related protein 1
- DUB
deubiquitinating enzyme
- GST
glutathione transferase
- Mfn
mitofusin
- MOM
mitochondrial outer membrane
- RNAi
RNA interference
- shRNA
short-hairpin RNA
- TM
transmembrane
- Ub
ubiquitin
- USP
ubiquitin-specific protease.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1103) on February 20, 2008.
REFERENCES
- Ahting U., Waizenegger T., Neupert W., Rapaport D. Signal-anchored proteins follow a unique insertion pathway into the outer membrane of mitochondria. J. Biol. Chem. 2005;280:48–53. doi: 10.1074/jbc.M410905200. [DOI] [PubMed] [Google Scholar]
- Altmann K., Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 2005;16:5410–5417. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik A. Y., Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Amerik A. Y., Nowak J., Swaminathan S., Hochstrasser M. The Doa4 deubiquitinating enzyme is functionally linked to the vacuolar protein-sorting and endocytic pathways. Mol. Biol. Cell. 2000;11:3365–3380. doi: 10.1091/mbc.11.10.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W., McCaffery J. M., King E. J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J. M. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Cerveny K. L., Tamura Y., Zhang Z., Jensen R. E., Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S. A., Ewald A. J., Griffin E. E., Fraser S. E., Chan D. C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detmer S. A., Chan D. C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Escobar-Henriques M., Westermann B., Langer T. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 2006;173:645–650. doi: 10.1083/jcb.200512079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Yokota S., Mihara K. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 2003;134:333–344. doi: 10.1093/jb/mvg150. [DOI] [PubMed] [Google Scholar]
- Fisk H. A., Yaffe M. P. A role for ubiquitination in mitochondrial inheritance in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:1199–1208. doi: 10.1083/jcb.145.6.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E. S., Leitner W. W., Robert E. G., Catez F., Smith C. L., Youle R. J. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- Gorsich S. W., Shaw J. M. Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell. 2004;15:4369–4381. doi: 10.1091/mbc.E03-12-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., van der Wel N. N., Orozco I. J., Peters P. J., van der Bliek A. M. Loss of the intermembrane space protein Mgm1/Opa1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 2004;279:18792–18798. doi: 10.1074/jbc.M400920200. [DOI] [PubMed] [Google Scholar]
- Hales K. G., Fuller M. T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., Berger S. L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G. J., Thatcher J. W., Mills J. P., Hales K. G., Fuller M. T., Nunnari J., Shaw J. M. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S., Aihara T., Hontani M., Okubo K., Hirose S. Mutational analysis of action of mitochondrial fusion factor mitofusin-2. J. Cell Sci. 2005;118:3153–3161. doi: 10.1242/jcs.02449. [DOI] [PubMed] [Google Scholar]
- Honda S., Hirose S. Stage-specific enhanced expression of mitochondrial fusion and fission factors during spermatogenesis in rat testis. Biochem. Biophys. Res. Commun. 2003;311:424–432. doi: 10.1016/j.bbrc.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- James D. I., Parone P. A., Mattenberger Y., Martinou J. C. hFis1, a novel component of the mammalian mitochondrial fission machinery. J. Biol. Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Kanaji S., Iwahashi J., Kida Y., Sakaguchi M., Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M., Neutzner A., Youle R. J. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner A., Kölling R. The yeast deubiquitinating enzyme Ubp16 is anchored to the outer mitochondrial membrane. FEBS Lett. 2003;549:135–140. doi: 10.1016/s0014-5793(03)00801-9. [DOI] [PubMed] [Google Scholar]
- Kovalenko A., Chable-Bessia C., Cantarella G., Israel A., Wallach D., Courtois G. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Labrousse A. M., Zappaterra M. D., Rube D. A., van der Bliek A. M. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Lee Y. J., Jeong S. Y., Karbowski M., Smith C. L., Youle R. J. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol. Biol. Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno E., Iura T., Mukai A., Yoshimori T., Kitamura N., Komada M. Regulation of epidermal growth factor receptor down-regulation by UBPY-mediated deubiquitination at endosomes. Mol. Biol. Cell. 2005;16:5163–5174. doi: 10.1091/mbc.E05-06-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Mozdy A. D., McCaffery J. M., Shaw J. M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Fukuda H., Kato A., Hirose S. MARCH-II is a syntaxin-6–binding protein involved in endosomal trafficking. Mol. Biol. Cell. 2005;16:1696–1710. doi: 10.1091/mbc.E04-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Kimura Y., Tokuda M., Honda S., Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naviglio S., Mattecucci C., Matoskova B., Nagase T., Nomura N., Di Fiore P. P., Draetta G. F. UBPY: a growth-regulated human ubiquitin isopeptidase. EMBO J. 1998;17:3241–3250. doi: 10.1093/emboj/17.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutzner A., Youle R. J. Instability of the mitofusin Fzo1 regulates mitochondrial morphology during the mating response of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:18598–18603. doi: 10.1074/jbc.M500807200. [DOI] [PubMed] [Google Scholar]
- Nijman S. M., Huang T. T., Dirac A. M., Brummelkamp T. R., Kerkhoven R. M., D'Andrea A. D., Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005a;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005b;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Nunnari J., Marshall W. F., Straight A., Murray A., Sedat J. W., Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol. Biol. Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 2003;278:7743–7746. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- Otsuga D., Keegan B. R., Brisch E., Thatcher J. W., Hermann G. J., Bleazard W., Shaw J. M. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa F. R., Hochstrasser M. The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature. 1993;366:313–319. doi: 10.1038/366313a0. [DOI] [PubMed] [Google Scholar]
- Parone P. A., James D. I., Da Cruz S., Mattenberger Y., Donzé O., Barja F., Martinou J. C. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis. Mol. Cell. Biol. 2006;26:7397–7408. doi: 10.1128/MCB.02282-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M., Fushman D. Polyubiquitin chains: polymeric protein signals. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Quesada V., Diaz-Perales A., Gutierrez-Fernandez A., Garabaya C., Cal S., Lopez-Otin C. Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem. Biophys. Res. Commun. 2004;314:54–62. doi: 10.1016/j.bbrc.2003.12.050. [DOI] [PubMed] [Google Scholar]
- Rapaport D. Finding the right organelle. Targeting signals in mitochondrial outer-membrane proteins. EMBO Rep. 2003;4:948–952. doi: 10.1038/sj.embor.embor937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:20150–20155. doi: 10.1074/jbc.273.32.20150. [DOI] [PubMed] [Google Scholar]
- Rinaldi T., Pick E., Gambadoro A., Zilli S., Maytal-Kivity V., Frontali L., Glickman M. H. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem. J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi T., Ricci C., Porro D., Bolotin-Fukuhara M., Frontali L. A mutation in a novel yeast proteasomal gene, RPN11/MPR1, produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA, and an altered mitochondrial morphology. Mol. Biol. Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A., Frank S., Gaume B., Herrler M., Youle R. J., Fuller M. T. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 2003;116:2763–2774. doi: 10.1242/jcs.00479. [DOI] [PubMed] [Google Scholar]
- Santel A., Fuller M. T. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R. E. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard K. A., Yaffe M. P. The yeast dynamin-like protein, Mgm1p, functions on the mitochondrial outer membrane to mediate mitochondrial inheritance. J. Cell Biol. 1999;144:711–720. doi: 10.1083/jcb.144.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skulachev V. P. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem. Sci. 2001;26:23–29. doi: 10.1016/s0968-0004(00)01735-7. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Griparic L., Shurland D. L., van der Bliek A. M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanovski D., Koutsopoulos O. S., Okamoto K., Ryan M. T. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J. Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- Suzuki H., Maeda M., Mihara K. Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 2002;115:1895–1905. doi: 10.1242/jcs.115.9.1895. [DOI] [PubMed] [Google Scholar]
- Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Wong E. D., Wagner J. A., Gorsich S. W., McCaffery J. M., Shaw J. M., Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonashiro R., et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Krueger E. W., Oswald B. J., McNiven M. A. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol. Cell. Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.