Abstract
Formins are conserved actin nucleators which promote the assembly of actin filaments for the formation of diverse actin structures. In fission yeast Schizosaccharomyces pombe, the formin cdc12p is required specifically in assembly of the actin-based contractile ring during cytokinesis. Here, using a mutational analysis of cdc12p, we identify regions of cdc12p responsible for ring assembly and localization. Profilin-binding residues of the FH1 domain regulate actin assembly and processive barbed-end capping by the FH2 domain. Studies using photobleaching (FRAP) and sensitivity to latrunculin A treatment show that profilin binding modulates the rapid dynamics of actin and cdc12p within the ring in vivo. Visualized by functional GFP-fusion constructs expressed from the endogenous promoter, cdc12p appears in a small number of cytoplasmic motile spot structures that deliver the formin to the ring assembly site, without detectable formation of an intermediate band of “nodes.” The FH3/DID region directs interphase spot localization, while an N-terminal region and the FH1-FH2 domains of cdc12p can target its localization to the ring. Mutations in putative DID and DAD regions do not alter regulation, suggesting that cdc12p is not regulated by a canonical autoinhibition mechanism. Our findings provide insights into the regulation of formin activity and the mechanisms of contractile ring dynamics and assembly.
INTRODUCTION
Cytokinesis in animal and fungal cells depends on the assembly and constriction of an actomyosin contractile ring. In the fission yeast Schizosaccharomyces pombe, the contractile ring assembles at the onset of mitosis in a medial position dictated by the predivisional nucleus (Daga and Chang, 2005). After anaphase chromosome segregation, the ring constricts inwards as a septal wall forms behind it. After completion of the septum, the central septum layer is broken down releasing two daughter cells (Le Goff et al., 1999; Guertin et al., 2002; Sipiczki, 2007).
Formins are a conserved family of actin nucleating proteins, which have been implicated in the assembly of diverse actin structures including stress fibers, filopodia, and adherens junctions, as well as cleavage furrows and contractile rings (Wallar and Alberts, 2003; Balasubramanian et al., 2004; Zigmond, 2004; Goode and Eck, 2007). The formin cdc12p was first identified in genetic screens for cytokinesis mutants in fission yeast (Nurse et al., 1976; Chang et al., 1996; Balasubramanian et al., 1998). Cdc12p function is specifically required during cytokinesis: it is a component of the contractile ring and is required for continuous actin filament assembly within the actin ring (Chang et al., 1997; Pelham and Chang, 2002).
In general, formins are able to catalyze actin nucleation, accelerate barbed polymerization, and protect growing filaments from capping activity (reviewed in Kovar, 2006; Goode and Eck, 2007). The conserved FH2 domain dimerizes to form a donut-shaped catalytic core that encircles the actin filament (Xu et al., 2004; Otomo et al., 2005). Through this domain, formins can maintain processive association with growing filament barbed ends and prevent binding by conventional capping proteins (Zigmond et al., 2003; Higashida et al., 2004; Moseley et al., 2004; Kovar and Pollard, 2004).
The activity of the FH2 domain is modulated by the proline-rich FH1 domain, which lies adjacent to the FH2 and binds to profilin. Interactions between the FH2, FH1 and profilin–actin complexes can increase the rate of barbed elongation of formin-capped actin filaments (Romero et al., 2004; Kovar, 2006; Vavylonis et al., 2006). Interactions between cdc12-cdc12p and S. pombe profilin (cdc3p) have been demonstrated by both genetic and biochemical studies (Chang et al., 1997; Pelham and Chang, 2002). In vitro studies demonstrate the importance of profilin for cdc12p activity; in the absence of profilin, cdc12p tightly caps barbed ends and blocks F-actin polymerization at this end of the filament. However, the addition of profilin can relieve this capping effect, essentially “gating” the barbed end, allowing rapid elongation of cdc12p-capped filament barbed ends (Kovar et al., 2005, 2006).
For proper cellular morphogenesis, formins must be targeted and activated at the correct time and place in a dynamic manner. The mechanisms responsible for formin localization and regulation are not well understood. Cdc12p is predicted to be inactive during interphase and then, in early mitosis, activated and delivered to the cell equator to assemble actin filaments for contractile ring formation and maintenance (Chang, 1999; Pelham and Chang, 2002). Many formins are regulated by autoinhibition, which is dependent on the intramolecular binding between N-terminal DID/FH3 and C-terminal DAD domains. Release from the autoinhibited state can be triggered by Rho GTPase binding to formin DID/FH3 regions (Alberts, 2001; Li and Higgs, 2003; Otomo et al., 2005; Schonichen et al., 2006; Seth et al., 2006). It is unknown whether cdc12p is regulated in this manner or if any of the S. pombe Rho proteins regulate cdc12p activity (Arellano et al., 1999).
For ring formation, contractile ring proteins are delivered to the future division site, where they assemble into a discrete ring structure. It has been proposed that cdc12p moves to the future division site in the form of a “cdc12p spot” or, alternatively, binds to multiple “nodes” on the equatorial plasma membrane (Chang, 1999; Wu and Pollard, 2005; Wu et al., 2006).
Here, we pursue a structure–function analysis of cdc12p to understand the mechanisms responsible for its function, localization, and regulation during cytokinesis. As the proline-rich FH1 domain has never been subject to in vivo mutational analysis in any formin, we analyzed the effects of specific mutations within this domain. We find that cdc12p is dynamic in the ring and that profilin binding increases the dynamic turnover of both actin and formin within the ring. In addition, we provide evidence that cdc12p exists in interphase spots and does not form cortical nodes during ring assembly. We also define multiple domains responsible for targeting cdc12p to different structures in distinct phases of the cell cycle. Our findings describing this cytokinesis-specific formin provide important new insights into formin function as well as the general mechanics of cell division.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetic Methods
S. pombe strains and oligonucleotides used in this study are listed in Supplementary Table 1. Standard methods were used for S. pombe for cell culture, cell staining, and molecular and genetic manipulations, as described at http://www.sanger.ac.uk/PostGenomics/S_pombe/docs/nurse_lab_manual.pdf.Yeast plasmid transformations were performed using the Frozen EZ Yeast Transformation Kit (Zymo Research, Orange, CA). Construction of yeast strains FC777 (cdc12::kanMX/cdc12+) and AY99 were performed by a PCR-based approach (Bahler et al., 1998). Linear PCR products generated from pFA6-KanMX deletion plasmid with oligonucleotides cdc12DF and cdc12DR or from pFA6-3GFP-KanMX (http://www-rcf.usc.edu/∼forsburg/pombeweb.html; Wu et al., 2003) with oligonucleotides cdc12CF and cdc12CR (Supplementary Table 1). PCR products were transformed into the cdc12::ura4/cdc12+ diploid strain FC777 (Chang et al., 1997), screened for G418 resistance, ura−, and confirmed by PCR.
Complementation Assays
Strain FC20 carrying mutant cdc12, wild-type cdc12+, or empty vector controls were grown to midlog phase in selective media, and 10-fold serial dilutions were spotted onto plates containing thiamine and incubated at 25 or 36°C for 2–4 d. Strain FC777 was transformed with all plasmids and allowed to sporulate. Spores were then plated on selective media and replica-plated onto YE5S medium containing 0.1 mg/ml G418 (Geneticin; Invitrogen, Rockville, MD) and scored for G418 resistance.
Plasmid Construction
Site-directed mutations were generated using Transformer Site-Directed Mutagenesis Kit (Clontech, Palo Alto, CA) with oligonucleotides listed in Supplementary Table 1. The cdc12 fragments used in plasmid constructions were derived from pFC103 and pnmt*-cdc12 (Supplementary Table 1). nmt* represents the medium strength version of the thiamine-regulated (41x) nmt1 promoter. pRL21 was constructed from pnmt*-cdc12 by removing the EcoRI site in the polylinker. pRL23 (pnmt*-cdc12-GFP) was constructed as follows: the C-terminal half of cdc12 (from the EcoRI site at 3135 base pairs of the cdc12 ORF to the PstI site in the polylinker of pFC103) was ligated into EcoRI and PstI sites in the polylinker of pUC19. Two BglII sites in this region of cdc12 were mutated to conservative changes (pRL16), and a novel BglII site was inserted immediately before the cdc12 stop codon (pRL17). The BamHI-BglII fragment of pFA6a-GFP-kanMX6 (Bahler et al., 1998) was inserted into this novel BglII site (pRL18). This C-terminal fragment of cdc12 fused to green fluorescent protein (GFP) coding sequences was exchanged with the EcoRI-XhoI fragment of pRL21 to make pRL23.
Mutations in the N-terminal half of cdc12 were generated in a 3.1-kb EcoRI cdc12 fragment cloned into pUC19. In cdc12-ΔN (pRL56), an EcoRI site was introduced at bp 526, and the N-terminus was deleted between the two EcoRI sites. Mutations in the C-terminal half of cdc12 were generated in pRL18. Oligonucleotides DAD-A1, DAD-A2, and DAD-A3 and their reverse complements were used sequentially to mutagenize pRL18 using Stratagene XL Mutagenesis Kit (La Jolla, CA), and EcoRI-BglII fragments were isolated and ligated into EcoRI-BglII sites of pRL23, replacing the wild-type cdc12 C-terminus and GFP coding sequences. To generate plasmids pRL86, pRL87, and pRL88, cdc12 fragments were amplified by PCR using the following oligonucleotides: RepXhoFH1 and FH2XhoGFPrev for pRL86, RepXhoFH1 and CtXhoGFPrev for pRL87, and RepXhoFH3, and FH2XhoGFPrev for pRL88 (Supplementary Table 1). The fragments were cotransformed with pREP42-EGFP-C (www-rcf.usc.edu/∼forsburg/vectors.html), linearized with BamHI for recombination in yeast. Plasmids pAY11-23 were generated by amplifying cdc12 fragments from pFC103, digesting PCR products containing NdeI and BamHI sites at their N- and C- termini, respectively, and ligating into NdeI and BamHI sites within pREP42-EGFP-C. Ligations were transformed into bacteria and amplified before transformation into yeast. For PCR, oligonucleotide primers N1 and N2 were used to generate the cdc12 fragment of pAY11, N1 and N5 for pAY12, N4 and N3 for pAY14, FH31 and FH32 for pAY15, N1 and FH32 for pAY16, SBD3 and SBD4 for pAY17, SBD3 and FH22 for pAY19, FH11 and FH22 for pAY20, C3 and C4 for pAY22, FH11 and C4 for pAY23, and N1 and FH22 for pAY25. All plasmid constructs were confirmed by DNA sequencing.
For in vitro actin assembly assays, wild-type His6-cdc12 FH1FH2p fragment (aa 870-1454) and mutant cdc12-ΔPBD FH1FH2p mutant fragment (aa 870-1454, Δ909-912, Δ942-953) were amplified from pFC103 with BamHI and PstI site overhangs, digested, and ligated into the expression vector pQE9 (Qiagen, Valencia, CA).
Microscopy and Pharmacological Inhibitors
Microscopy was performed using a wide-field fluorescence microscope or a spinning disk confocal microscope using Open Lab software (Improvision, Lexington, MA) for image acquisition and analysis, as described (Pelham and Chang, 2001). Cells for imaging were grown overnight to midlog phase and imaged on pads made from 1% agarose in media. For visualization of cdc12 mutant and partial fragment GFP constructs, cells that were maintained in the presence of thiamine were cultured in thiamine-free media for 18–20 h to induce expression from the pnmt* promoter and were imaged live. F-actin was visualized with either Alexa568- or 488-labeled phalloidin (Molecular Probes, Eugene, OR), and staining was performed using 300s permeabilization in 1% Triton and a 15-min fixation period in 4% formaldehyde. For Alexa568 staining, cells were rinsed once with buffer before imaging. Latrunculin A (LatA; 100 μM, Sigma-Aldrich, St. Louis, MO) in DMSO was used for actin depolymerization experiments.
Fluorescence recovery after photobleaching (FRAP) studies were performed using a Zeiss LSM510 Meta scanning confocal microscope (Thornwood, NY) with an Ar/He laser. Images were acquired at 10-s intervals using 10% laser power at 488 nm; photobleaching was performed at maximum laser power by 30× iterative scanning focused on a circular region of 1-μm diameter around one edge of a cytokinetic ring. Images were analyzed using ImageJ software (http://rsb.info.nih.gov/ij/). Kymographs were constructed for each cell from image slices encompassing the ring. Mean fluorescence intensity was measured using a line scan analysis function within the bleached area. Background subtraction was done using fluorescence values outside the cell, and values were corrected for photobleaching by dividing by the fraction of initial intensity within an adjacent unbleached cell. Values were normalized to set starting fluorescence intensity equal to 1 and postbleach values to 0. Data for each strain were grouped and averaged.
Profilin-binding Assays
His6-tagged cdc12p fragment containing the FH1 domain (aa 763-1091) was bacterially expressed and purified from pFC122 as described (Chang et al., 1997). His6-tagged FH1 mutants were amplified by PCR from full-length mutant constructs and used to replace the wild-type FH1 fragment in pFC122. Resulting protein products were then expressed and purified in a similar manner. Glutathione S-transferase (GST)-Cdc3p (profilin) was bacterially expressed from pKG251 (Balasubramanian et al., 1994). Proteins were expressed in Escherichia coli (BL21; Stratagene), affinity-purified on nickel (Qiagen), or glutathione (Invitrogen, Carlsbad, CA) agarose beads. Cdc12p protein fragments were purified in 8 M urea-PBS, renatured by dialysis into PBS, and precleared before binding assays by microcentrifugation at 14,000 × g for 10 min at 4°C. For in vitro binding experiments, purified His-tagged cdc12 proteins were incubated with GST-profilin–bound or GST control glutathione beads for 1 h in 200 μl CB buffer (0.6 M sorbitol, 50 mM Tris, pH 7.5, 140 mM NaCl, 5 mM EDTA, 0.06% Triton X-100) before centrifugation to separate supernatant and pellet samples as described in Chang et al. (1997).
Actin Filament Assembly and Elongation Assays
His6-fusion proteins were expressed in E. coli (BL21; Stratagene) and purified on nickel resin (Qiagen). After elution with 300 mM imidazole and subsequent dialysis in HEK buffer (20 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl), proteins were purified further by anion exchange chromatography (Mono-Q; Amersham Biosciences, Piscataway, NJ) and gel filtration chromatography (Superose 12; Amersham Biosciences). Rabbit skeletal muscle actin was purified as described by Spudich and Watt (1971) and labeled with pyrenyliodoacetamide as described (Pollard, 1984; Higgs et al., 1999). Monomeric actin was prepared by gel filtration using a Sephacryl S-200 column (Amersham Biosciences) equilibrated in G-buffer (10 mM Tris, pH 8.0, 0.2 mM ATP, 0.2 mM CaCl2, 0.2 mM DTT). Untagged S. pombe profilin (cdc3p) was expressed in E. coli using expression vector pMW172 (Eads et al., 1998; Lu and Pollard, 2001) and purified by anion exchange chromatography (HiTrap-Q and Mono-Q; Amersham Biosciences) followed by gel filtration (Superdex75; Amersham Biosciences).
Actin filament assembly assays were performed as described (Moseley et al., 2006) using gel filtered G-actin. Actin (final concentration 4 μM, 5% pyrene-labeled) was mixed with 15 μl of HEK buffer or proteins in HEK buffer (250 nM cdc12 FH1FH2p or cdc12 ΔPBD FH1FH2p, and/or 10 μM cdc3p), and then reaction components (57 μl) were mixed immediately with 3 μl of 20× initiation mix (40 mM MgCl2, 10 mM ATP, 1 M KCl) to initiate actin assembly. Pyrene fluorescence was monitored at 365-nm excitation and 407-nm emission in a fluorescence spectrophotometer (Photon Technology International, Lawrenceville, NJ) at 25°C. Filament elongation assays were performed as described (Sagot et al., 2002; Moseley et al., 2006) using 75 nM wild-type cdc12 FH1FH2p or cdc12-ΔPBD FH1FH2p and 2 μM cdc3p.
RESULTS
FH1 and FH2 Domains Are Necessary But Not Sufficient for cdc12p Function
To determine the function of specific protein domains in cdc12p function, we constructed a series of defined deletions and substitutions in cdc12 by site-directed mutagenesis (Figures 1 and 2A). These constructs were expressed from a thiamine-regulated nmt1* (41× medium strength) promoter on multicopy plasmids as C-terminal GFP fusion proteins to facilitate protein localization studies. The presence of a C-terminal GFP tag does not perturb protein function, as replacement of cdc12+ with cdc12-GFP at the chromosomal locus produces no measurable cell growth or cytokinesis defect (Chang, 1999).
Figure 1.
cdc12 mutant constructs. cdc12 constructs were generated by site-directed mutagenesis and expressed as C-terminally tagged GFP fusions under the control of the medium strength nmt* (41x) promoter on Rep-based plasmids. (A and B) Diagram of the mutant constructs. The cdc12 FH3 region (aa 321-503) overlaps with sequences (aa 486-535) homologous to the mDia1 DID domain (see Supplementary Figure 3). The M region contains putative dimerization (DD; aa 550-630) and coiled coil (CC; aa 680-714 and 863-883) domains. The mutant protein FH2A represents an FH2 domain substitution of the conserved residues (GNYMND) to alanines. (C) Function of mutant cdc12 proteins, as assessed by their ability to rescue growth of temperature-sensitive cdc12-112 mutant cells at 36°C or of cdc12Δ mutants in the presence of thiamine.
Figure 2.
FH1 domain mutations disrupt profilin binding. (A) Mutations in the FH1 domain. Amino acid sequences of the wild-type and mutant (ΔPP1, ΔPP2, ΔPBD, and L913P, ΔPP2) FH1 polyproline repeats PP1 (blue) and PP2 (green) are shown. Deleted or mutated residues are shown in red. (B) Coomassie-stained gel of bacterially expressed and purified proteins used in profilin pulldowns. (C) Cdc12p FH1 pulldown assay: wild-type, ΔPBD, and L913P, ΔPP2 protein fragments and GST-cdc3p (or GST alone) were incubated together with glutathione beads before pelleting. Total (t), pellet (p), and supernatant (s) protein fractions were analyzed by SDS-PAGE. (D) cdc12 FH1 mutants localize to the ring. Images of cdc12+-GFP, and ΔPP1 or ΔPP2 cdc12-GFP mutants expressed in cdc12Δ cells, and of ΔPBD and L913P cdc12-GFP mutants expressed in a cdc12+ background. Scale bar, 5 μm.
We assayed these cdc12 mutant genes for their ability to rescue growth in temperature-sensitive cdc12-112 and cdc12 null mutant backgrounds. As expected, the conserved catalytic FH1 and FH2 domains were essential for cdc12+ function. Specific mutations in the FH1 and FH2 in the context of the whole cdc12 protein abolished the ability to rescue either cdc12-ts or cdc12 null strains. Alanine substitutions of six highly conserved residues (GNYMND) in the FH2 domain that reside in a predicted key α-helix within the dimerization “post” structure (Xu et al., 2004; Higgs and Peterson, 2005), rendered the cdc12-ΔFH2A mutant protein nonfunctional (Figure 1 and Figure S1). However, the FH1-FH2 domains were clearly not sufficient for full cdc12+ function in cytokinesis. A cdc12p fragment containing only the catalytic FH1-FH2 domains did not rescue a cdc12 null strain.
No single region outside the FH1-FH2 domain was strictly required for cdc12p function. Deletions of other single domains did not abolish cdc12 function, as these alleles could still drive actin ring assembly and sustain cell viability (Figure 1 and Figure S1 and S2). Truncated cdc12 mutant proteins lacking the entire N-terminal region of cdc12p upstream of the FH1-FH2 domain (FH1FH2-C) or mutant proteins bearing large deletions spanning the C-terminus (ΔC1 and ΔC2) were functional. Thus, for rescue, portions of either the N- or C-terminal regions were needed in addition to the catalytic FH1-FH2 domains.
For complementation of the temperature-sensitive cdc12-112 strain, only the FH1 and FH2 domains were required (Figure 1). The cdc12-112 mutant still expresses a full-length protein (data not shown), so this “mutant protein” may be able to function with FH1-FH2p fragments at a low level to support viability.
The cdc12 FH1 Domain Has Two Functionally Redundant Profilin-binding Sites
Although it has been established that formins interact with profilin through the FH1 domain polyproline repeats (Chang et al., 1997; Watanabe et al., 1997; Ozaki-Kuroda et al., 2001; Sagot et al., 2002; Kovar et al., 2003), the function of these interactions has not been well characterized in vivo. Mutations in the polyproline-binding site of the fission yeast profilin cdc3p show defects in cdc12p regulation and cytokinesis (Lu and Pollard, 2001; Kovar et al., 2003); however, it is difficult to rule out that these mutations may affect the many activities of profilin in unpredictable ways. No directed mutational analyses of proline-rich sequences within the FH1 have been reported in any formin. In contrast to some FH1 domains in mammalian formins, which contain large number of proline-rich tracts (Kovar, 2006), the cdc12 FH1 domain contains only two stretches of consecutive proline residues, which we named PP1 and PP2. The PP1, like many formin polyproline motifs, consists of a string of proline residues interrupted by a hydrophobic residue at the penultimate position (Figure 2A; Frazier and Field, 1997; Mahoney et al., 1997, 1999).
We created defined mutations in these proline-rich regions: polyproline repeats were deleted separately (ΔPP1 and ΔPP2) or together (ΔPBD; Figure 2A). We confirmed that these polyproline sites are necessary for binding to profilin. Using in vitro assays with bacterially expressed proteins, we demonstrated that the ΔPBD mutant FH1 fragment no longer bound to profilin (Figure 2C). FH1 proteins with only a single polyproline region deleted (ΔPP1 and ΔPP2) were still competent to bind profilin (our unpublished observations). To test the importance of the penultimate leucine residue in PP1, we replaced this leucine in PP1 with another proline and deleted PP2 (L913P, ΔPP2; Figure 2A). The L913P, ΔPP2 FH1 protein fragment was also deficient in profilin binding (Figure 2), demonstrating that residues in addition to the prolines influence the affinity of these sites for profilin.
To determine whether the polyproline tracts were necessary for function in vivo, these FH1 mutations were introduced into the full-length cdc12 gene. Deletion of both polyproline sites abolished cdc12p function in complementation assays (Figure 1 and Figure S1 and S2). Mutant proteins bearing deletions of either single polyproline tract (ΔPP1 and ΔPP2) were functional and produced no apparent defect in cytokinesis. The mutant in the penultimate leucine of PP1 (L913P, ΔPP2) was not functional (Figure S1 and S2). When expressed in a wild-type background, these FH1 mutant proteins displayed normal localization patterns, suggesting that these mutations did not grossly affect expression or protein localization (Figure 2D). These data indicate that profilin binding is critical for cdc12p function in vivo and that the two polyproline tracts are functionally redundant.
FH1 Domain Profilin-binding Drives Actin Assembly and Inhibits Actin Capping
We next addressed how profilin-binding defects in the ΔPBD mutant affect actin assembly by cdc12p. Previous work has shown that upon filament nucleation, cdc12p tightly caps fast-growing barbed ends and thus restricts filament growth to pointed ends. However, profilin-cdc12p interactions release or “gate” this capping and permit barbed-end growth of cdc12p-associated filaments (Kovar et al., 2003, 2006). In actin assembly assays, we confirmed that purified wild-type cdc12 FH1FH2p nucleates filament assembly. The mutant cdc12 ΔPBD FH1FH2p, bearing the profilin-binding site double deletion, stimulated actin assembly with kinetics similar to that of the wild-type fragment (Figure 3B). These results, which are consistent with previous findings using profilin mutants defective in polyproline-binding and wild-type cdc12p fragment (Kovar et al., 2003), show that profilin binding is not required for nucleation of actin by cdc12p.
Figure 3.
Activities of a cdc12p profilin-binding mutant in vitro. (A) Coomassie-stained SDS-PAGE gel of purified wild-type and ΔPBD mutant FH1FH2 proteins. (B) The effects of cdc12p wild-type or ΔPBD His6 -FH1FH2 fusion proteins and S. pombe profilin (cdc3p) on actin polymerization, using a pyrene actin assembly assay. Actin (4 μM; 5% pyrene-labeled) was incubated with indicated concentrations of either wild-type or ΔPBD cdc12 FH1FH2p and/or cdc3p. F-actin accumulation was represented by the increase in pyrene fluorescence (arbitrary units) over time (s). (C) Filament elongation assay. Effects of wild-type and ΔPBD cdc12 FH1FH2 proteins on barbed-end elongation of pre-existing actin filaments in the presence or absence of profilin. Monomeric actin (0.5 μM, 10% pyrene labeled) was polymerized at the barbed ends of mechanically sheared actin filament seeds (333 nM) in the presence of indicated concentrations of cdc12 FH1FH2p and/or cdc3p.
Addition of purified S. pombe profilin (cdc3p) to wild-type cdc12 FH1FH2p enhanced both the rate and extent of filament formation, likely by “gating” cdc12p-capping activity and permitting barbed-end growth, as seen previously (Figure 3B; Kovar et al., 2003, 2006). In parallel assays with the cdc12 profilin binding mutant fragment (ΔPBD FH1-FH2p), addition of profilin failed to promote growth and, instead, completely inhibited actin polymerization (Figure 3B). These results suggest that profilin binding by FH1 is required for filament elongation by cdc12p.
To test the role of the FH1 in “gating” of cdc12p barbed-end capping, we assayed rates of barbed-end elongation from preformed F-actin seeds. In the absence of profilin, both wild-type and ΔPBD FH1FH2p blocked barbed-end assembly (Figure 3C), consistent with tight capping activity (Kovar et al., 2003, 2005). In our assay, the addition of profilin to wild-type FH1FH2p was sufficient to gate cdc12p activity and permit barbed-end polymerization at 70% of the rate of free barbed ends. In contrast, addition of profilin to reactions with ΔPBD FH1FH2p failed to release capping activity and had no effect on barbed-end polymerization (Figure 3C). Similar to the behavior of wild-type cdc12p in the absence of profilin, the loss of profilin binding activity of the FH1 domain led to constitutive, profilin-insensitive filament capping by cdc12p in vitro.
Profilin Binding Promotes cdc12p and Actin Dynamics in Vivo
We next examined whether profilin binding affects the dynamics of actin and cdc12p in contractile rings in vivo. Studies have revealed that the contractile ring is a dynamic structure in many cell types (Pelham and Chang, 2002; Hotulainen et al., 2005; Murthy and Wadsworth, 2005) In fission yeast, actin, tropomyosin, myosin II, and myosin light chains have been shown to have turnover periods of less than 1 min (Pelham and Chang, 2002; Wong et al., 2002). Whether cdc12p is equally dynamic or resides as a stable nucleating machine at the cell cortex was not known. To measure the dynamics of cdc12p within the ring, we conducted FRAP studies in which cdc12-GFP was photobleached on one side of the ring, and fluorescence recovery was monitored over time (Figure 4, A and B). In cells mildly overexpressing cdc12-GFP, the formin displayed rapid recovery with an average t1/2 = 30 s (Figure 4, C and D). Because cdc12p is not an abundant ring component (Wu and Pollard, 2005), moderate overexpression facilitated robust measurements over time and also allowed direct comparison with the behavior of nonfunctional mutant cdc12 proteins. We observed identical recovery kinetics in cells expressing endogenous levels of cdc12-3GFP expressed from the chromosomal locus (t1/2 = 32 s; data not shown), indicating that overexpression had no significant effect cdc12p turnover. Thus, cdc12p in the contractile ring is highly dynamic and exhibits turnover rates similar to that of both F-actin and myosin.
Figure 4.
Profilin binding modulates dynamics of cdc12p and actin in the ring. FRAP of wild-type cdc12-GFP and ΔPBD cdc12-GFP within contractile rings expressed in wild-type cells. Cells were imaged in a single medial Z-plane, in which the ring appears as two dots on either side of the cell. (A) Cdc12-GFP was photobleached on one side of the contractile ring (yellow circle) at t = 0 and imaged at 10-s intervals in a single focal plane through the middle of cell using a scanning confocal microscope. Scale bar,1 μm. (B) Kymographs of image slices encompassing the ring (green box in A) over time. Scale bar, 1 μm. (C) Average fluorescence intensities in the photobleached area over time after photobleaching. Values were normalized to initial fluorescence intensity (100% at t = −10 s) and after bleach (0% at t = 0). (D) Mean fluorescence recovery periods. Error bars, SDs. p < 0.01 between WT and mutant recovery times. cdc12+, n = 12; ΔPP1, n = 7; and ΔPBD, n = 10. (E) Cells expressing wild-type cdc12-GFP (top panels) and ΔPBD cdc12-GFP (bottom panels) were treated with 100 μM LatA for indicated time periods, fixed, and stained with Alexa546-phalloidin. Arrows show actin rings that persist in cdc12ΔPBD cells longer than in wild-type cells. (F) Persistence of actin rings in presence of 100 μM LatA (n > 1000 cells scored for each allele). Scale bar, 5 μm.
We then measured the dynamics of the profilin-binding mutant cdc12 ΔPBD-GFP. These experiments bear the caveat that we had to examine the dynamics of this tagged mutant protein expressed in a cdc12+ background, because cells expressing cdc12 ΔPBDp alone are not viable. Fluorescence recovery of cdc12 ΔPBD-GFP had an average recovery t1/2 = 66 s, twofold slower than wild-type cdc12-GFP under the same conditions (Figures 4, B–D). cdc12-ΔPP1-GFP displayed an intermediate recovery rate of t1/2 = 49 s (Figures 4, C and D). A similar trend was also seen in the magnitude of recovery after bleaching, suggesting a greater subset of stable molecules in the mutant alleles. Thus, profilin binding appears to increase the turnover dynamics of cdc12p in the contractile ring.
Next, we examined whether profilin binding affects the dynamics of actin filaments in vivo. Although no functional fluorescent fusion proteins with actin have yet been developed, actin dynamics can be probed by measuring the rate of actin depolymerization in cells treated with the actin monomer-sequestering molecule LatA. When cdc12+ cells expressing wild-type cdc12-GFP were treated with LatA, 90% of actin rings depolymerized within 80 s of drug addition (n > 1000 cells; Figure 4, E and F; Pelham and Chang, 2002). Given its capping activities in vitro, we hypothesized that cdc12 ΔPBDp protein might stabilize actin filaments at the ring. Indeed, cells expressing the cdc12 ΔPBD-GFP mutant protein retained rings three times longer after LatA treatment; 90% of actin rings were depolymerized only after 4 min of drug exposure (n > 1000 cells). Consistent with the specificity of cdc12p function in the contractile ring (Pelham and Chang, 2002), cdc12 ΔPBD-GFP had no stabilizing effect on other actin structures such as actin patches or cables (Figure 4E).
In summary, these in vivo and in vitro studies show that profilin binding at the FH1 domain regulates the ability of cdc12p to elongate and cap actin filaments. In vivo, cdc12p-profilin interactions modulate the dynamics of actin filaments and cdc12p at the ring.
Localization of cdc12p to Interphase Spots
We next sought to determine the domains of cdc12p responsible for its localization. First, however, we re-examined the localization behavior of cdc12p throughout the cell cycle. cdc12p and cdc12-GFP have been shown to form a cytoplasmic spot structure during interphase upon overexpression (Chang et al., 1997, 1999). This spot exhibits both actin- and microtubule-based motility, moves to the equatorial region, and then extends into the contractile ring structure during ring assembly in early mitosis. Immunofluorescence of wild-type cells using an anti-cdc12p antibody further supported the existence of such a spot (Chang, 1999). However, cdc12p interphase spot structures are not readily apparent in cells expressing a cdc12 fusion with a single GFP from the chromosomal locus (data not shown).
Recent studies (Wu et al., 2006; Vavylonis et al., 2008) have questioned the existence of such an interphase spot structure, proposing instead that the ring arises from a broad medial band of node structures containing cdc12p, myosin, and other ring proteins. Before ring formation, myosin II (myo2p) and its light chains localizes to a wide punctate band of more than 50 nodes, which slowly coalesce into a tight ring structure (Naqvi et al., 1999; Motegi et al., 2000; Wu et al., 2003). However, the presence of cdc12p in these nodal structures has not clearly been documented in wild-type cells under normal conditions. The images of cdc12p published in Wu et al. (2006) are more consistent with cdc12p localization to “loose” rings during initial ring formation or to abnormal multiple rings (in cdc25-22 mutants), which later merge into a single ring structure (Daga and Chang, 2005). A distinct pattern of cdc12p in multiple medial nodes is only apparent after prolonged treatment with LatA (Wu et al., 2006; our unpublished observations); we note that this LatA-induced localization pattern does not necessarily reflect a normal intermediate in ring assembly.
As the distribution of cdc12p is critical to the understanding the events in initial ring assembly, we tried to resolve these different models of cdc12p localization by generating an integrated cdc12-triple tandem GFP strain. This cdc12-3GFP protein is expressed from the endogenous cdc12 promoter at the native chromosomal locus and is functional, as determined by its ability to promote normal cytokinesis and ring assembly as the sole copy of cdc12 protein in the cell (data not shown).
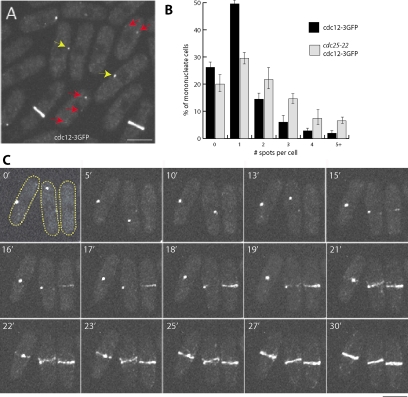
Confocal microscopy revealed that cdc12-3GFP localized to motile cytoplasmic spots in interphase cells (Figure 5A). Although the spots were often dim, the majority (74%) of interphase cells (cells with one nucleus but no ring) contained detectable cdc12p spots. Most of these cells possessed a single large spot, whereas a subset exhibited multiple (n = 2–5) smaller spots (Figure 5B; n > 1000 cells). Small cdc12p spots were sometimes observed to merge together into a larger spot (data not shown). At the onset of mitosis, we observed cdc12p spots at the medial cortex rapidly spreading out in a linear manner to encircle the cell circumference, forming the ring (Figure 5C; Supplementary Movie 1). All spots were seen to incorporate into the ring (100% spots, n = 78). In the minority of cells with no detectable spot, rings appeared to form without any spot or node intermediate. We never observed the formation of an intermediate broad band consisting of numerous (e.g., 50+) cdc12p “nodes,” as proposed by Wu et al. (2006). We saw the same behavior in cdc12-3YFP strains used by Wu et al., 2006 (data not shown). cdc25 strains often exhibited slightly more, smaller spots, but their numbers never approached 50+ nodes. The observed behavior of interphase spots were consistent with our previous findings with overexpressed cdc12p proteins (Chang et al., 1997, 1999). Thus, our results show that during initial phases of ring formation, cdc12p localizes to one or a small number of interphase spots that spread laterally into a ring.
Figure 5.
Cdc12p interphase spots give rise to mitotic rings. (A and C) Cells expressing endogenous levels of a functional cdc12-3GFP fusion were grown to exponential phase in YE5S at 25°C and imaged using spinning disk confocal microscopy. Maximum intensity projections of 3D sections are shown. Yellow arrows show cells with a single dominant interphase cdc12p spot; red arrows mark cells with multiple smaller cdc12p spots. (B) Number of cdc12-3GFP spots detected in interphase wild-type and cdc25-22 backgrounds at 25°C. (n > 1000) (C) Time-lapse images of three cells expressing cdc12–3GFP (two with single interphase spots, one without); rapid ring formation occurs without detectable node intermediates. Scale bars, 5 μm.
An N-terminal Region Directs cdc12p Localization to Interphase Spots
To determine which region(s) of cdc12p mediates its localization behavior, we examined the cellular distribution of cdc12 fragments tagged with GFP (Figures 1 and 6). These constructs were imaged in wild-type backgrounds, and those constructs that could support viability were also examined in a cdc12 null background.
Figure 6.
Localization of cdc12 mutant proteins. Wild-type and mutant cdc12-GFP proteins were expressed from a medium strength nmt* promoter on a REP41x-based plasmid in cells grown in the absence of thiamine for 16–20 h. Functional constructs (see Figure 1) were expressed and imaged in a cdc12::kanMX null background. Similar results were found these constructs were expressed in cdc12+ cells (data not shown). Nonfunctional mutants and small fragments were expressed in a cdc12+ background. (A) Images of cdc12-GFP fusion protein localizations. Maximum intensity projections of 3D stacks are shown. Arrow highlights N-FH3-GFP construct forming an interphase spot structure. (B) Schematic summary of the ability of constructs to localize to mitotic rings or interphase spots. Scale bar, 5 μm.
An N-terminal region was required specifically for localization to interphase spot structures. The FH3 region was originally designated as a region of homology among formins (Petersen et al., 1998). Subsequent structural analyses of mDia1 showed that this region contains distinct structural domains including the diaphanous inhibitory domain (DID; Higgs and Peterson, 2005; Otomo et al., 2005; Goode and Eck, 2007). cdc12p contains an N-terminal region with weak similarity to the DID domain; the FH3 covers part of the predicted DID domain, but not the dimerization domain (DD) or coiled coil (CC) domains (Figure 1; Supplementary Figure 3). A construct cdc12 ΔFH3-GFP, in which the FH3 (including a portion of the predicted DID region) was deleted, did not localize to spots during interphase, but was still targeted to rings in mitotic cells, even when expressed in a cdc12Δ background. Conversely, as previously shown (Petersen et al., 1998), an FH3-GFP protein localized to spot-like structures, but not to mitotic rings (Figure 6; data not shown). Nuclear localization was also noted. Thus, this N-terminal region is necessary and sufficient for cdc12p localization to interphase spots.
The ΔFH3 mutant allele allowed us to examine the functional significance of cdc12p spot localization. Cells expressing cdc12 ΔFH3-GFP as the sole copy of cdc12p assembled functional actin rings in the absence of interphase spot formation (Figure 6; data not shown). Like wild-type cdc12p, these cdc12 ΔFH3-GFP rings assembled without the appearance of an intermediate broad band structure. Time-lapse images showed formation of a clear ring from no defined structure within 3 min (Supplementary Movie 2). Thus, ring assembly can occur in the absence of detectable cdc12p spot formation.
Multiple Elements Target cdc12p to Mitotic Rings
Other regions of cdc12p were found to target it to the ring in mitosis. We found that N-terminal fragments of the protein (N and N151) was sufficient for ring localization (Figure 6). Truncation of the first 50 residues of the N-terminal fragment (ΔN50) abolished targeting activity. This suggests the presence of a ring localization element at within the first 151 residues of cdc12p, in a region distinct from the FH3/DID domain. Interestingly, a fragment containing both the N-terminus and the FH3/DID domain was able to recapitulate both spot and ring localization patterns (Figure 6).
However, this N-terminal region was not solely responsible for ring localization. When the N-terminal 151 aa were deleted in the context of the whole protein, the resulting mutant cdc12 ΔN-GFP still localized normally to rings and spots, even when expressed in a cdc12Δ background (Figure 6). Thus, other region(s) within cdc12p must also mediate targeting to mitotic rings.
One additional region may be the FH1-FH2 domain. An FH1-FH2 fragment localized weakly to actin rings when expressed in wild-type cells (Figure 6). This ring localization was apparent in cells expressing relatively low levels of this fragment. At higher expression levels, FH1-FH2p generated abnormal linear or punctate aggregates containing F-actin (Figure 6; data not shown). The FH1-FH2 fragment may localize to the ring by binding actin filament barbed ends and/or through hetero-dimerization with endogenous cdc12p in these cells.
The addition of the C-terminal region, which improves the functionality of the FH1-FH2 domain (Figure 1 and Figure S1), also improved its ring localization. When expressed in a wild-type background, an FH1FH2-Cp fragment localized more robustly to the division site than the FH1-FH2 alone. However, it also generated abnormal rings and other cytoplasmic structures, such as abnormal aster-like structures, which contained the cdc12 fragment and F-actin, at the division plane (Figure 6; data not shown). We were unable to test the targeting of the C-terminal region alone, as expression of this fragment localized to the nucleus (data not shown). Thus, cdc12p can be targeted to the ring through multiple independent elements.
Testing the Function of a Putative cdc12p Diaphanous Autoregulatory Domain
The activity and localization of many formins are regulated by an autoinhibitory mechanism which is dependent on the intramolecular binding between DID and diaphanous autoregulatory domain (DAD) domains in the N- and C-terminal regions of the formin, respectively (Alberts, 2001; Higgs, 2005; Higgs and Peterson, 2005; Rivero et al., 2005; Seth et al., 2006). DAD domains are loosely conserved motifs with a core consensus (MDXLLXXL), often closely followed by a cluster of basic residues (Li and Higgs, 2005; Nezami et al., 2006; Wallar et al., 2006). Whether cdc12p is regulated by such a mechanism is not known, but it does harbor a potential DAD domain sequence just downstream of its catalytic FH2 domain (Figure 7A; Higgs and Peterson, 2005).
Figure 7.
Characterization of mutants in a putative cdc12p DAD domain. (A) Alignment of a cdc12 DAD-like domain with DADs of Diaphanous family formins (Higgs, 2005). (B) Mutations in the putative cdc12 DAD domain. (C) Cells expressing cdc12 DAD-A3-GFP mutant from a REP42x plasmid vector. GFP localization and phalloidin staining showed normal cdc12p and F-actin distributions. Scale bar, 5 μm.
We generated alanine substitution mutations at the most conserved residues within the putative cdc12 DAD consensus motif (Figure 7B). Cells expressing the mutant cdc12 DAD-GFP fusion proteins exhibited no strong cytokinesis phenotype and did not appear to have an obvious increase in general actin filament assembly (Figure 7C). In addition, cells expressing cdc12 ΔFH3p, in which the DID domain was disrupted, also displayed no obvious effects on cdc12p function (Figures S1 and S2; data not shown).
We tested a model in which a conformational change dependent on the putative DAD and DID domains drives the transition from cdc12p spot structure to a ring in early mitosis. Although cdc12 ΔFH3p mutants did not form spots, the cdc12 DAD mutant proteins localized normally to interphase spots and mitotic rings (Figure 7C). Thus, these results suggest that a DAD-dependent autoinhibitory mechanism is not responsible for promoting spot formation. Further, the effects of disrupting the putative DID domain were not solely due to loss of a putative DAD interaction. Thus, cdc12p may be regulated by alternative or additional modes.
DISCUSSION
Here, we have examined the functions of cdc12p domains in contractile ring assembly and localization. Although the FH2 domain comprises the catalytic core of the formin, other domains contribute multiple activities to target cdc12p properly within the cell and modulate actin assembly and dynamics. The proline-rich FH1 domain regulates the behavior of the FH2 domain on actin filament barbed ends and influences actin dynamics and stability within the contractile ring. Multiple sites mediate the localization of cdc12p to the mitotic ring, whereas the FH3/DID region governs localization to interphase spots. Our findings on the dynamics of cdc12p provide new insights into the mechanism of ring assembly and actin ring structure during cytokinesis.
Cdc12p, Profilin, and Dynamic Actin Filament Assembly
Mutational analyses of the FH1 domain show that critical residues lie in two conserved and functionally redundant polyproline tracts. Although formin FH1 domains generally have multiple binding sites for profilin, cdc12p function in vivo requires only one polyproline tract that binds to profilin. A conserved hydrophobic residue near the end of the polyproline tract is necessary for profilin binding and function, perhaps by helping to orient profilin binding on the polyproline tract (Mahoney et al., 1999).
Our findings add to a growing collection of genetic and biochemical evidence that formin interactions with profilin are required for their interdependent functions in cytokinesis and other actin-based cellular processes. In vitro studies suggest that profilin may act as a cofactor for cdc12p to stimulate barbed-end elongation by increasing the local concentration of profilin-bound G-actin in the vicinity of filament barbed ends attached to the FH2 domain (Kovar, 2006). Profilin also provides a “gating” function at the barbed end of the actin filament (Figure 3; Kovar et al., 2003). Our studies provide important evidence that the FH1 polyproline domains provide these essential functions both in vitro and in vivo.
In vivo, the interaction of cdc12p formin with profilin regulates ring dynamics. There is growing appreciation that the mature contractile ring is a dynamic structure, with individual molecules exchanging on the order of 1 min. FRAP shows that cdc12p molecules associate transiently with the ring with turnover rates similar to both F-actin and myosin (Pelham and Chang, 2002; Wong et al., 2002). Comparable dynamics have been observed for S. pombe for3p and Saccharomyces cerevisiae Bni1p at sites of polarized cell growth, but not S. cerevisiae Bnr1p, which is much more stably bound at the bud neck (Martin and Chang, 2006; Buttery et al., 2007).
As with yeast actin cables, the contractile ring is composed of short filaments assembled from sites on the plasma membrane (Kamasaki et al., 2007). Analyses of the actin cable–generating formins (for3p and Bni1p) suggest that these formins bind transiently to the cortex, before being released on the barbed ends of short actin filaments within a cable (Martin and Chang, 2006; Buttery et al., 2007). We speculate that the transient nature of cdc12p localization may reflect similar cortical docking and release events of cdc12p in the contractile ring. The profilin-binding mutants of cdc12p can stabilize actin and their own association with the ring in a dominant manner. This coupling of dynamics suggests that these mutant proteins stabilize actin in the ring by capping barbed ends. It is possible that cdc12p can also modulate processive filament depolymerization in a manner dependent on profilin binding. We show here that formin and profilin not only regulate actin filament assembly, but can also modulate filament stability.
Ring Assembly and the cdc12p Spot
A recently proposed model for contractile ring assembly is based on the observation that myosin and other ring factors are initially targeted to the medial cortex in a mid1p-dependent manner into a punctate band of “nodes,” which then slowly move together into a ring structure using forces generated by actin–myosin interactions (Wu et al., 2006; Vavylonis et al., 2008). A critical element of this model is the assumption that cdc12p resides in these nodes to mediate actin assembly from these nodes. Here, we show that cdc12p is not detectable in a large number of these nodes but, rather, moves to the cell division plane in the form of a single or small number of particles. Although initially observed in cells overexpressing cdc12p, we confirmed the existence of cdc12p spots in interphase cells when cdc12p is expressed from its endogenous chromosomal promoter. Time-lapse imaging shows that a single or small number (≤5) of cdc12p spots dock at the equatorial region and then quickly spread into well-defined rings. No clear intermediate localization to nodes was ever observed under our sensitive imaging conditions, which can detect very small numbers of molecules (≥3 GFPs; Martin and Chang, 2006). Our studies cannot, however, rule out the presence of highly transient, motile cdc12p molecules in nodes that are not detectable by fluorescence microscopy. The spot-based model is supported further by recent electron micrographs showing that the barbed ends of actin filaments in the ring are not randomly distributed during early mitosis, but can be predominantly polarized toward a common point, suggesting a focused origin of nucleation (Kamasaki et al., 2007).
The cdc12p spot structure, however, is not essential for ring formation. Spots are not seen in a cdc12 ΔFH3 mutant, yet well-defined, functional actin rings are still assembled without nodal intermediate structures. Further studies will examine whether spot formation facilitates formin delivery to the site of ring assembly or influences the organization of actin filaments in the ring.
The FH3 region may function in spot formation by interacting with other proteins or by affecting intramolecular interactions. One candidate interacting protein is cdc15p (PCH protein), which may colocalize with cdc12p in both interphase spots and rings, and binds directly to an N-terminal region of cdc12p that includes the FH3/DID region (aa 1-524; Carnahan and Gould, 2003). Another possibility is that the DID-like sequences of cdc12p mediate a conformational change responsible for spot formation through the intramolecular binding between DID and DAD domains (Higgs and Peterson, 2005; Nezami et al., 2006); however, this model is inconsistent with the finding that cdc12 DAD mutants still form interphase spots. This FH3/DID region is required for the localization of other formins; for example, S. pombe fus1p to mating projection tips, S. cerevisiae Bni1 to bud necks, and human mDia1 to mitotic spindles (Petersen et al., 1998; Kato et al., 2001; Ozaki-Kuroda et al., 2001).
A combination of interactions may mediate targeting of cdc12 formin to actin rings. Ring localization was not perturbed by deletions of any single region of cdc12p. The extreme N-terminus contains a domain that is sufficient for ring localization. It is not clear if cdc15p mediates this localization, as the region responsible for cdc15p binding (Carnahan and Gould, 2003) differs from the boundaries we defined for ring localization. N-terminal localization domains have been mapped in other formins (Kato et al., 2001; Evangelista et al., 2002; Nakano et al., 2002; Takeya and Sumimoto, 2003; Martin et al., 2005), but no clearly conserved motifs are apparent in this region. No regulation by Rho GTPases has been demonstrated for cdc12p, and so it is not clear whether this region could mediate Rho binding. Similarly, multiple sites in the S. pombe formin for3p are responsible for targeting this formin to cell tips (Martin et al., 2007). Further studies defining the mechanisms of regulation and localization for this cytokinesis-specific formin will be critical for understanding fundamental mechanisms underlying cytokinesis.
Supplementary Material
ACKNOWLEDGMENTS
We thank Z. Perlman, N. Padte, S. Bratman, N. Minc, M. Ellis, H. Niederstrasser, R. Basu, S. Salas-Pino, R. Daga, M. Shirasu-Hiza, C. Field, B. Feierbach, A. Paoletti, S. Almo, J. Q. Wu, (The Ohio State University), T. Pollard, and J. Pringle for discussions, technical support, and comments on the manuscript and A. Manning, (Dartmouth College), J. Q. Wu, D. Kovar, (University of Chicago), and T. Pollard (Yale University) for generously sharing strains and biochemical reagents. FRAP studies were performed at the Columbia Confocal Facility. Funding was provided by National Institutes of Health (NIH) training grant DK07328 to A.Y., NIH Grant R01/GM63691 to B.L.G., and NIH Grant R01/GM56836, and grants from American Cancer Society, Hirschl Charitable Trust, and March of Dimes Foundation to F.C.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0731) on February 27, 2008.
REFERENCES
- Alberts A. S. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Arellano M., Coll P. M., Perez P. Rho GTPases in the control of cell morphology, cell polarity, and actin localization in fission yeast. Microsc. Res. Tech. 1999;47:51–60. doi: 10.1002/(SICI)1097-0029(19991001)47:1<51::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., Bi E., Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M. K., Hirani B. R., Burke J. D., Gould K. L. The Schizosaccharomyces pombe cdc3+ gene encodes a profilin essential for cytokinesis. J. Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M. K., McCollum D., Chang L., Wong K. C., Naqvi N. I., He X., Sazer S., Gould K. L. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery S. M., Yoshida S., Pellman D. Yeast formins Bni1 and Bnr1 utilize different modes of cortical interaction during the assembly of actin cables. Mol. Biol. Cell. 2007;18:1826–1838. doi: 10.1091/mbc.E06-09-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan R. H., Gould K. L. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J. Cell Biol. 2003;162:851–862. doi: 10.1083/jcb.200305012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. Movement of a cytokinesis factor cdc12p to the site of cell division. Curr. Biol. 1999;9:849–852. doi: 10.1016/s0960-9822(99)80372-8. [DOI] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 1997;137:169–182. doi: 10.1083/jcb.137.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Woollard A., Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 1996;109(Pt 1):131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Daga R. R., Chang F. Dynamic positioning of the fission yeast cell division plane. Proc. Natl. Acad. Sci. USA. 2005;102:8228–8232. doi: 10.1073/pnas.0409021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads J. C., Mahoney N. M., Vorobiev S., Bresnick A. R., Wen K. K., Rubenstein P. A., Haarer B. K., Almo S. C. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- Evangelista M., Pruyne D., Amberg D. C., Boone C., Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat. Cell Biol. 2002;4:260–269. doi: 10.1038/ncb770. [DOI] [PubMed] [Google Scholar]
- Frazier J. A., Field C. M. Actin cytoskeleton: are FH proteins local organizers? Curr. Biol. 1997;7:R414–R417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- Goode B. L., Eck M. J. Mechanism and function of formins in control of actin assembly. Annu. Rev. Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Guertin D. A., Trautmann S., McCollum D. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashida C., Miyoshi T., Fujita A., Oceguera-Yanez F., Monypenny J., Andou Y., Narumiya S., Watanabe N. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 2004;303:2007–2010. doi: 10.1126/science.1093923. [DOI] [PubMed] [Google Scholar]
- Higgs H. N. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Higgs H. N., Blanchoin L., Pollard T. D. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Higgs H. N., Peterson K. J. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P., Paunola E., Vartiainen M. K., Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol. Biol. Cell. 2005;16:649–664. doi: 10.1091/mbc.E04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T., Osumi M., Mabuchi I. Three-D arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Watanabe N., Morishima Y., Fujita A., Ishizaki T., Narumiya S. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J. Cell Sci. 2001;114:775–784. doi: 10.1242/jcs.114.4.775. [DOI] [PubMed] [Google Scholar]
- Kovar D. R. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Kuhn J. R., Tichy A. L., Pollard T. D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003;161:875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Pollard T. D. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Wu J. Q., Pollard T. D. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell. 2005;16:2313–2324. doi: 10.1091/mbc.E04-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X., Utzig S., Simanis V. Controlling septation in fission yeast: finding the middle, and timing it right. Curr. Genet. 1999;35:571–584. doi: 10.1007/s002940050455. [DOI] [PubMed] [Google Scholar]
- Li F., Higgs H. N. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Li F., Higgs H. N. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- Lu J., Pollard T. D. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol. Biol. Cell. 2001;12:1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney N. M., Janmey P. A., Almo S. C. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat. Struct. Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- Mahoney N. M., Rozwarski D. A., Fedorov E., Fedorov A. A., Almo S. C. Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat. Struct. Biol. 1999;6:666–671. doi: 10.1038/10722. [DOI] [PubMed] [Google Scholar]
- Martin S. G., Chang F. Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Martin S. G., McDonald W. H., Yates J. R., 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Martin S. G., Rincon S. A., Basu R., Perez P., Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. B., Maiti S., Goode B. L. Formin proteins: purification and measurement of effects on actin assembly. Methods Enzymol. 2006;406:215–234. doi: 10.1016/S0076-6879(06)06016-2. [DOI] [PubMed] [Google Scholar]
- Moseley J. B., Sagot I., Manning A. L., Xu Y., Eck M. J., Pellman D., Goode B. L. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi F., Nakano K., Mabuchi I. Molecular mechanism of myosin–II assembly at the division site in Schizosaccharomyces pombe. J. Cell Sci. 2000;113(Pt 10):1813–1825. doi: 10.1242/jcs.113.10.1813. [DOI] [PubMed] [Google Scholar]
- Murthy K., Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Nakano K., Imai J., Arai R., Toh E. A., Matsui Y., Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- Naqvi N. I., Eng K., Gould K. L., Balasubramanian M. K. Evidence for F-actin-dependent and -independent mechanisms involved in assembly and stability of the medial actomyosin ring in fission yeast. EMBO J. 1999;18:854–862. doi: 10.1093/emboj/18.4.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezami A. G., Poy F., Eck M. J. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Nurse P., Thuriaux P., Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Otomo T., Otomo C., Tomchick D. R., Machius M., Rosen M. K. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda K., Yamamoto Y., Nohara H., Kinoshita M., Fujiwara T., Irie K., Takai Y. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R. J., Chang F. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat. Cell Biol. 2001;3:235–244. doi: 10.1038/35060020. [DOI] [PubMed] [Google Scholar]
- Pelham R. J., Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Petersen J., Nielsen O., Egel R., Hagan I. M. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. Polymerization of ADP-actin. J. Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero F., Muramoto T., Meyer A. K., Urushihara H., Uyeda T. Q., Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6:28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M. F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sagot I., Rodal A. A., Moseley J., Goode B. L., Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Schonichen A., Alexander M., Gasteier J. E., Cuesta F. E., Fackler O. T., Geyer M. Biochemical characterization of the diaphanous autoregulatory interaction in the formin homology protein FHOD1. J. Biol. Chem. 2006;281:5084–5093. doi: 10.1074/jbc.M509226200. [DOI] [PubMed] [Google Scholar]
- Seth A., Otomo C., Rosen M. K. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J. Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Splitting of the fission yeast septum. FEMS Yeast Res. 2007;7:761–770. doi: 10.1111/j.1567-1364.2007.00266.x. [DOI] [PubMed] [Google Scholar]
- Spudich J. A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Takeya R., Sumimoto H. Fhos, a mammalian formin, directly binds to F-actin via a region N-terminal to the FH1 domain and forms a homotypic complex via the FH2 domain to promote actin fiber formation. J. Cell Sci. 2003;116:4567–4575. doi: 10.1242/jcs.00769. [DOI] [PubMed] [Google Scholar]
- Vavylonis D., Kovar D. R., O'Shaughnessy B., Pollard T. D. Model of formin-associated actin filament elongation. Mol. Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D., Wu J. Q., Hao S., O'Shaughnessy B., Pollard T. D. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wallar B. J., Alberts A. S. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 2003;13:435–446. doi: 10.1016/s0962-8924(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Wallar B. J., Stropich B. N., Schoenherr J. A., Holman H. A., Kitchen S. M., Alberts A. S. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J. Biol. Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B. M., Narumiya S. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 1997;16:3044–3056. doi: 10.1093/emboj/16.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K. C., D'Souza V., M., Naqvi N. I., Motegi F., Mabuchi I., Balasubramanian M. K. Importance of a myosin II-containing progenitor for actomyosin ring assembly in fission yeast. Curr. Biol. 2002;12:724–729. doi: 10.1016/s0960-9822(02)00790-x. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Kuhn J. R., Kovar D. R., Pollard T. D. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev. Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Pollard T. D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Sirotkin V., Kovar D. R., Lord M., Beltzner C. C., Kuhn J. R., Pollard T. D. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Zigmond S. H. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004;16:99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Zigmond S. H., Evangelista M., Boone C., Yang C., Dar A. C., Sicheri F., Forkey J., Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.