Abstract
N3-methyl-adenine (3MeA) is the major cytotoxic lesion formed in DNA by SN2 methylating agents. The lesion presumably blocks progression of cellular replicases because the N3-methyl group hinders interactions between the polymerase and the minor groove of DNA. However, this hypothesis has yet to be rigorously proven, as 3MeA is intrinsically unstable and is converted to an abasic site, which itself is a blocking lesion. To circumvent these problems, we have chemically synthesized a 3-deaza analog of 3MeA (3dMeA) as a stable phosphoramidite and have incorporated the analog into synthetic oligonucleotides that have been used in vitro as templates for DNA replication. As expected, the 3dMeA lesion blocked both human DNA polymerases α and δ. In contrast, human polymerases η, ι and κ, as well as Saccharomyces cerevisiae polη were able to bypass the lesion, albeit with varying efficiencies and accuracy. To confirm the physiological relevance of our findings, we show that in S. cerevisiae lacking Mag1-dependent 3MeA repair, polη (Rad30) contributes to the survival of cells exposed to methyl methanesulfonate (MMS) and in the absence of Mag1, Rad30 and Rev3, human polymerases η, ι and κ are capable of restoring MMS-resistance to the normally MMS-sensitive strain.
INTRODUCTION
DNA is subject to a variety of chemical modifications that alter its structure. Such alterations can block basic cellular functions such as transcription and/or replication and can lead to cell death, mutagenesis and cancer in higher eukaryotes. One such modification is DNA methylation, which can be caused by endogenous chemicals, products of metabolism, environmental exposure or treatment with several cancer chemotherapeutics. Not surprisingly, cells have developed several evolutionarily conserved mechanisms for repairing or tolerating this type of DNA damage, including base excision repair (BER), nucleotide excision repair (NER), recombination and translesion DNA synthesis (TLS) (1).
Methylating agents primarily react with exocylic nitrogen or oxygen atoms on purines and pyrmidines, with the reaction mechanism (SN1 or SN2) determining the relative ratio of oxygen to nitrogen modifications (2). The major products in DNA exposed to SN2 methylating agents are N7-methylguanine and N3-methyladenine (3MeA), while there is very little methylation of oxygen atoms on the bases or the sugar phosphate backbone. 3MeA accounts for ∼20% of the base damage formed by SN2 methylating agents (2) and is considered to be the major cytotoxic lesion produced by such chemicals, based on the fact that bacterial and viral DNA polymerases are blocked before adenine residues but not guanine, on templates treated with either SN1 or SN2 methylating agents (3).
3MeA is primarily removed by BER, although NER appears to provide an important back-up mechanism in the absence of BER in eukaryotes (4–7). Mouse embryonic fibroblasts (MEFs) lacking Aag, the DNA glycosylase that normally removes 3MeA from DNA, are sensitive to methyl methanesulfonate (MMS) and the compound methyl lexitropsin, which preferentially methylates N3 of adenine (8). Indeed, Aag−/− cells become arrested in S phase longer than their wild-type counterparts treated with either methylating agent, suggesting that the unrepaired 3MeA residues are a block to replication in vivo. However, it has been extremely difficult to prove that 3MeA blocks replication directly, as the half-life of 3MeA in vitro is estimated to be between 12 and 24 h (9), thereby precluding biochemical analysis. Furthermore, assuming 3MeA has a similar, or even faster decay in vivo, it seems likely that by the time the MMS-treated Aag−/− cells arrest in S phase, a significant portion of the 3MeA residues would be converted to replication-blocking abasic sites. The fact that the arrested cells eventually complete S phase (8) suggests that the replication-block is either removed by another repair mechanism, or that specialized DNA polymerases are able to bypass the damaged site.
Several eukaryotic DNA polymerases are capable of performing TLS. Perhaps the best-characterized eukaryotic TLS polymerases are polζ, a B-family polymerase (10,11), and polη, polι, polκ and Rev1, all of which are Y-family polymerases (12). Based upon structural studies, the Y-family polymerases appear to be good candidates to facilitate TLS of 3MeA, since unlike high-fidelity replicative polymerases, they do not make the same contacts with N3 of adenine in the minor groove of duplex DNA (13).
A major obstacle that has to date prevented the study of 3MeA TLS in vitro has been the inherent instability of the 3MeA lesion. To circumvent these problems, we have synthesized a stable 3-deaza analog of the nucleoside 3-methyl-2′-deoxyadenosine that can be incorporated into synthetic oligonucleotides as 3-deaza-3-methyladenine (3dMeA). Here, we show that human replicative polymerases polα and polδ are blocked by 3dMeA, while human and Saccharomyces cerevisiae Y-family polymerases are capable of bypassing the modified base in vitro. In agreement with our in vitro observations, we also demonstrate that human DNA polymerases η, ι and κ have the ability to restore MMS-resistance to a normally MMS-sensitive mag1Δ rad30Δ rev3Δ strain of S. cerevisiae.
MATERIALS AND METHODS
Oligonucleotides
Ethenoadenosine phosphoramidite was purchased from Glen Research (Sterling, VA, USA). All oligonucleotides used for in vitro replication and PCR assays, were synthesized by Lofstrand Labs Limited (Gaithersburg, MD, USA) and gel purified prior to use. Ethenoadenine and 3dMeA bases were incorporated into oligonucleotides using ultra-mild synthesis conditions.
Enzymes
Human polδ (14), GST-polι (15), His-polη (16) and S. cerevisiae polζ (GST-Rev3/Rev7) (17),were purified as previously described. Human polα was purchased from Chimerx (Milwaukee, WI, USA). Human polκ, S. cerevisiae polη and Rev1 protein were purchased from Enzymax (Lexington, KY, USA). Mouse Aag was purchased from Trevigen (Gaithersburg, MD, USA).
Synthesis of the 3-deaza-3-methyl-dA-phosphoramidite
A detailed protocol outlining the chemical synthesis of the 3-deaza-3-methyl-dA-phosphoramidite is available online as Supplementary Data.
In vitro Aag excision assay
To measure DNA glycosylase activity on various substrates, 5′-[32P] 29mer, 5′-GCT CGT CAG ACG ATT TAG AGT CTG CAG TG-3′ (with the adenine, ethenoadenine or 3dMeA underlined and in bold font), was annealed to its complementary strand. Double-stranded DNA of 0.4 pmol was treated with 3 U of mAag or mock treated for 1 h at 37°C. NaOH was added to a final concentration of 100 mM along with 10 mM Tris, 1 mM EDTA (final) and the samples were incubated at 37°C to cleave any resulting abasic sites. Samples were resolved on a 15% gel (8-M urea) and visualized with a Molecular Dynamics phosphorimager and ImageQuant software.
Replication assays
In vitro replication assays were performed using the 29mer oligonucleotide 5′-GCT CGT CAG ACG ATT TAG AGT CTG CAG TG-3′ as a template (with the location of the undamaged adenine, or 3dMeA underlined and in bold font). For most experiments described herein, this template was annealed to a [32P]-labeled 16mer primer with the following sequence; 5′-CAC TGC AGA CTC TAA A -3′. For the extension assays reported in Table 3, the [32P]-labeled primer was a 17mer with the sequence; 5′-CAC TGC AGA CTC TAA AX -3′, where X is either A, or T. Primer-template DNAs were prepared by annealing the 5′ [32P]-labeled primer to the unlabeled template DNA at a molar ratio of 1:1.5. Standard 10-μl reactions contained 40 mM Tris–HCl at pH 8.0, 5 mM MgCl2, 100 μM of each ultrapure dNTP (Amersham Pharmacia Biotech, NJ, USA), 10 mM DTT, 250 μg/ml BSA, 2.5% glycerol and 10 nM primer/template DNA. The concentration of polymerase added varied and is given in the legends to figures 3, 4, 5 and 7. After incubation at 37°C (or 30°C for yeast enzymes) for 5 min, reactions were terminated by the addition of 10 μl of 95% formamide/10 mM EDTA and the samples heated to 100°C for 5 min and briefly chilled on ice. Reaction mixtures (5 μl) were resolved on 15% polyacrylamide, 8M urea gels and analyzed with a Molecular Dynamics phosphorimager and ImageQuant software.
Table 3.
Kinetics of single nucleotide extensiona from matched/mismatch primer termini paired with 3-deaza-3-methyl adenine (3dMeA) by Y-family DNA polymerases
| Polymeraseb | Primer: Template | Vmax (%ext/min) | Km (μM) | Vmax/Km (μM−1 min−1) | fext |
|---|---|---|---|---|---|
| Hs η | T:A | 1.1 | 0.6 | 1.8 | 1 |
| T:3dMeA | 1.1 | 7.7 | 0.14 | 8.0 × 10−2 | |
| A:3dMeA | 0.77 | 4.9 | 0.16 | 9.0 × 10−2 | |
| Sc η | T:A | 2.0 | 1.0 | 2 | 1 |
| T:3dMeA | 4.0 | 14.4 | 0.27 | 1.35 × 10−1 | |
| A:3dMeA | 1.14 | 18 | 0.06 | 3.0 × 10−2 | |
| Hs ιc | T:A | 7.8 | 1.1 | 7.1 | 1 |
| T:3dMeA | 1.8 | 6.4 | 0.28 | 4.0 × 10−2 | |
| A:3dMeA | 0.8 | 14 | 0.06 | 8.5 × 10−3 | |
| Hs κ | T:A | 1.0 | 1.4 | 0.7 | 1 |
| T:3dMeA | 0.63 | 9.4 | 0.07 | 1.0 × 10−1 | |
| A:3dMeA | 0.36 | 17.6 | 0.02 | 3.0 × 10−2 |
aIncorporation of C opposite undamaged G.
bHs, Homo sapien; Sc, S. cerevisiae.
cIn the presence of 0.25 mM MgCl2.
Steady-state reaction conditions
For steady-state kinetic reactions, each polymerase was assayed to determine the amount of enzyme and nucleotide that would result in <20% incorporation (18,19): 0.4 U/reaction for polα, 1.2 nM for human polη, 1.8 nM for polι, 1.5 nM for polκ and 1.4 nM for S. cerevisiae polη. All reactions were performed in 10 μl in the standard reaction buffer described earlier, except those involving polι, where the concentration of magnesium chloride was reduced from 5 to 0.25 mM. Reactions were initiated by the addition of the dNTP and lasted for 1.5–5 min for the correct nucleotides and 5–10 min for incorrect nucleotides, depending on the polymerase. On unmodified templates, dNTP concentrations ranged from 0.01 to 100 μM for the correct dTTP and from 1 to 500 μM for the incorrect dNTPs. For Y-family polymerases on the 3dMeA containing template, dTTP concentrations ranged from 0.1 μM to 1 mM while incorrect dNTPs ranged from 10 μM to 1 mM (except for polι reactions where dATP ranged from 2 to 100 μM, while dGTP and dCTP ranged from 10 to 300 μM). For the data shown in Table 3, dCTP concentrations varied from 0.2 to 10 μM on the undamaged template and from 10 to 300 μM for 3dMeA-containing template. For polα with the 3dMeA-containing template, dATP and dTTP were varied from 0.1 to 1 mM. Replication products were separated on 15% polyacrylamide gels containing 8-M urea and visualized with a Molecular Dynamics phosphorimager and quantified with ImageQuant software.
The apparent Vmax and Km values for each enzyme and nucleotide were determined from a Hanes–Woolf plot by linear least-squares fit as described previously (18). The catalytic efficiency of nucleotide insertion was calculated as the ration of Vmax/Km and the frequency of misinsertion was calculated as (Vmax/Km)incorrect/(Vmax/Km)correct as described previously (18) using SigmaPlot software (SPSS, Chicago, USA).
Generation of yeast strains and plasmids
All yeast strains were derived from the W303 background (20). MAG1 was disrupted by PCR amplification of the URA3 gene from pRS416 using primers with 40 nt of homology to upstream and downstream of MAG1 (MagUraF, 5′-ATG AAA CTA AAA AGG GAG TAT GAT GAG TTA ATA AAA GCA GCA GAG CAG ATT GTA CTG AGA GTG C-3′ and MagUraR, 5′-TTA GGA TTT CAC GAA ATT TTC TTC TGC CTT CAT CAT GGC AGC GGT ATT TTC TCC TTA CGC-3′) and transformed into C10-15a (W303 RAD5 + mata) (20). Positive disruptants were confirmed by PCR and MMS sensitivity. The mag1Δ haploid strain was mated to C10-10a, in order to obtain the mag1Δ rad30Δ double mutant (BPC1-4d) and a backcrossed mag1Δ (BPC1-2a) strain. BPC1-4d (mag1Δ::URA3 rad30Δ:HIS3 matα) was mated with C17-1A (rev3Δ:HisG-URA3 mata) to obtain mag1Δ rev3Δ (BPC2-8c), rad30Δ rev3Δ (BPC2-5a) double mutants and the mag1Δ rad30Δ rev3Δ (BPC2-13c) triple mutant. Since MAG1 and REV3 disruptions were both marked by the URA3 gene, all strains genotypes were confirmed by PCR for these two genes by triplex PCR with the following reverse primer for URA3 (URA3_44R; 5′-ACT AGG ATG AGT AGC AGC ACG-3′) and forward and reverse primers for either MAG1 (MAG1_95upF; 5′-TGG CCA CTG CCC TCT GAT ATG-3′ and MAG1_298R; 5′-CTT GGC CAC TGA TCT GTT GAG-3′) or REV3 (REV3_355upF; 5′-ACC ATT GTC CAA AGC TGT CGC-3′ and REV3_223R; 5′-ACG TGG CAC AAT ACT TGA TGC C-3′).
Plasmids expressing human and S. cerevisiae Y-family polymerases were constructed from pESC-LEU (Stratagene, La Jolla, CA, USA). POLI was cloned by digesting p6-1 (21) with NcoI, filling in the overhang with Klenow fragment, followed by digestion with AvaI and subsequent cloning into the SmaI site of pESC-LEU to generate pBP65. POLH was cloned as a NotI–BamHI fragment from pCDNA-XPV (22) into pESC-LEU digested with NotI and BglII to generate pPB66. POLK was cloned into pESC-LEU by first digesting pBP65 with NcoI, filling the ends with Klenow fragment to blunt end and subsequently digesting the vector with XmaI. An EcoRV–XmaI fragment from pHSE2 (a kind gift from Haruo Ohmori, University of Kyoto, Japan), encoding POLK was subsequently cloned into the vector to generate pBP98. Saccharomyces cerevisiae RAD30 was cloned as an NcoI–PstI fragment from pJM231 into the similarly digested plasmid, pBP65, to generate pBP82.
Survival assays
MMS toxicity for each genotype was assessed on overnight cultures. Yeast were harvested and washed twice with PBS. MMS was diluted to 0.25% in PBS and aliquots of each strain were removed at selected time intervals, washed with PBS and diluted for plating on YPAD agar plates. Colonies were counted after 5 days at 30°C. For the complementation assays, strain BPC2-13A (mag1Δ rad30Δ rev3Δ) was transformed with pBP65 (expresses human polι), pBP66 (expresses human polη), pBP98 (expresses human polκ), pBP82 (expresses S. cerevisiae polη) or pESC-LEU. Yeast strains were cultured overnight in complete synthetic raffinose medium lacking l-leucine. One hour prior to MMS treatment, the cultures were harvested by centrifugation and transferred to synthetic galactose medium to induce the expression of polymerases. Cells were harvested and treated as described above, except dilutions of each culture were plated on synthetic galactose agar plates lacking l-leucine.
RESULTS
3dMeA is a stable analog of N3-methyladenine
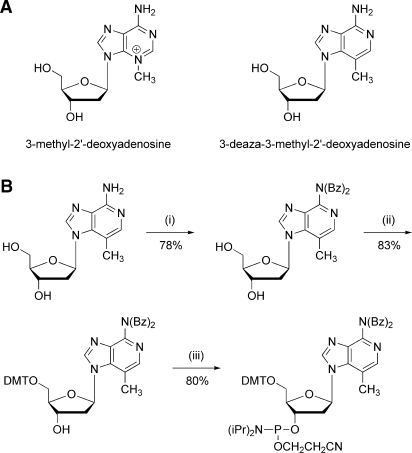
3-Methyl adenosine is unstable in vitro with an estimated half-life of just 12–24 h (23). This short half-life has therefore limited biochemical or enzymatic studies on the lesion. To circumvent these problems, we have synthesized a 3-deaza-3-methyl-2′-deoxyadenosine analog of 3-methyl-2′-deoxyadenosine. The 3-deaza- analog has the same overall structure as the naturally occurring adduct (Figure 1A), but it lacks the positive charge associated with the N3 atom that normally destabilizes the glycosidic bond, and is therefore very stable. The analog can be synthesized as a phosphoramidite (Figure 1B) and can be incorporated into oligonucleotides by standard chemical DNA synthesis.
Figure 1.
Synthesis of a synthetic 3-deaza-3-methyl-dA phosphoramidite. (A) Chemical structures of 3-methyl-2′-deoxyadenosine and 3-deaza-3-methyl-2′-deoxyadenosine. Replacement of the N3 with carbon removed the positive charge and helps stabilize the glycosidic bond. (B) Schematic of the synthesis of the 3-deaza-3-methyl-dA phosphoramidite (i) Di-benzoylation of 3-deaza-dA using benzoyl chloride in pyridine. (ii) Dimethoxytritylation using dimethoxytrityl chloride in pyridine. (iii) Phosphitylation using N,N-diisopropylamino-(2-cyanoethyl)phosphoramidic chloride and diisopropylethylamine in dichloromethane.
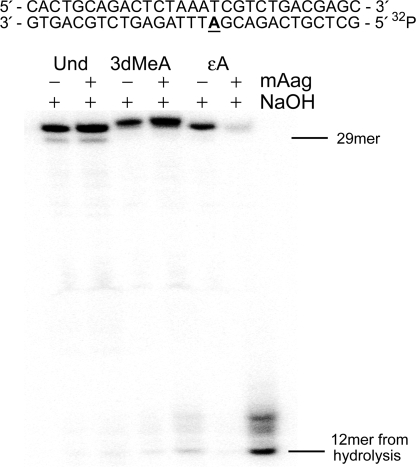
Since 3MeA is excised from DNA by the alkyladenine DNA glycosylase (Aag) (24), we determined if 3dMeA is also a substrate for Aag by treating either unmodified duplex DNA or DNA containing 3dMeA, or ethenoadenine (εA) with purified mouse Aag followed by hydroxide treatment. εA is a well-characterized substrate for Aag (25) and as noted in Figure 2, is completely excised from the substrate, as all of the εA oligonucleotide is cleaved at the resulting abasic site, by hydroxide treatment (Figure 2). In contrast, the 3dMeA containing DNA shows relatively little cleaved substrate, and there is no detectable cleavage product in the unmodified control. This demonstrates that Aag can excise 3dMeA, but to a much lesser extent than εA and presumably the naturally occurring 3MeA.
Figure 2.
3-deaza-3-methyl adenine is a stable analog of 3MeA. Mouse alkyladenine glycosylase (mAag) excises both 3-methyladenine (3MeA) and ethenodeoxyA (εA). 0.4 pmol of undamaged, 3dMeA- or εA-containing DNA was treated with 3 U of mAag, or mock treated for 1 h at 37°C. To hydrolyze the resulting abasic sites, NaOH was added to a final concentration of 100 mM along with 10 mM Tris, 1 mM EDTA (final) and the samples were incubated at 37°C. Samples were resolved on a 15% polyacrylamide gel containing 8-M urea. The nucleotide sequence of the 29mer duplex DNA is shown at the top of the panel and the position of the uncleaved 32P-labeled 29mer oligonucleotide and 12mer product are shown on the right side of the gel.
Treatment with NaOH in the absence of Aag confirms that 3dMeA analog is indeed stable and that even boiling of the DNA to anneal the lesion containing strand to its complementary strand, did not result in abasic sites that could be subsequently hydrolyzed by treatment with NaOH. We suspect that the 3dMeA analog may not be removed as readily as naturally occurring 3MeA because of its stabilized glycosidic bond. Indeed, it has been proposed that the weakened glycosidic bond of several Aag substrates may facilitate excision by the glycosylase (26).
3dMeA is a strong kinetic block to replicative polymerases, but is bypassed by Y-family polymerases
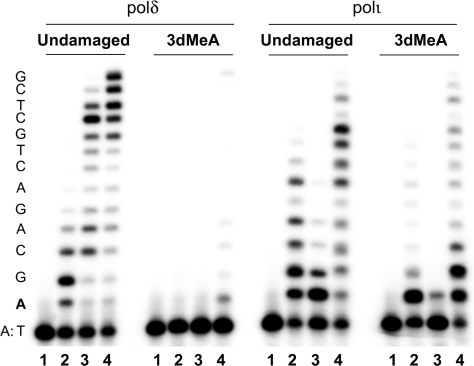
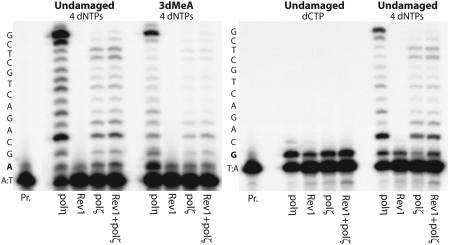
To date, there has been no direct evidence of 3MeA blocking a replicative DNA polymerase. We therefore compared human polα and polδ to polη, polι, and polκ in the presence of the four standard deoxynucleotides to determine which enzymes were capable of replicating a template containing 3dMeA. Under standard reaction conditions and with an undamaged template, each polymerases utilizes ∼10–20% of the primer (Figure 3A, left), however, virtually no extension of the primer annealed to the 3dMeA-containing template was observed in the presence of polα and polδ, indicating that the lesion is a strong kinetic block to replicative polymerases. By comparison, both incorporation and bypass of the lesion was observed in the presence of human polη, ι or κ (Figure 3). Steady-state kinetic analyses revealed that incorporation of T opposite the 3dMeA lesion only occurred with an efficiency of 0.15–3% of that opposite an undamaged A (Table 1). However, when one compares the catalytic activity (Vmax/Km) of the Y-family enzymes ability to incorporate opposite the 3dMeA lesion, it is 125- to 1200-fold more efficient than the incorporation by polα (Table 1) (full kinetic parameters are supplied as Supplementary Data).
Figure 3.
Ability of human DNA polymerases to bypass 3-deaza-3-methyl adenine in vitro. Standard reactions contained 100 μM all 4 dNTPs and lasted for 5 min at 37°C. Reactions contained 0.2 U polα, 5 nM polδ, 3 nM polη, 1.5 nM polι and 3 nM polκ. The nucleotide sequence of the template DNA is shown on the left-hand side of the gel. The ‘A’ in bold font is either undamaged (left-hand panel) or 3-deaza-3-methyl adenine (3dMeA; right-hand panel). As clearly seen, the 3dMeA lesion is a strong block to replication by human DNA polymerases α and δ, but can be bypassed by human polymerases η, ι and κ.
Table 1.
Efficiency of insertion of T opposite undamaged A, or 3dMeA by various eukaryotic polymerases
| Polymerasea | Template | Vmax/Km (μM−1 min−1) | Template | Vmax/Km (μM−1 min−1) | Efficiency of insertionb | Efficiency of insertionc |
|---|---|---|---|---|---|---|
| Hs α | A | 3.4 | 3dMeA | 0.0002 | 5.88 × 10−5 | 1 |
| Hs η | A | 3.06 | 3dMeA | 0.073 | 2.30 × 10−2 | 365 |
| Hs ιd | A | 210 | 3dMeA | 0.24 | 1.14 × 10−3 | 1200 |
| Hs κ | A | 13.73 | 3dMeA | 0.025 | 1.82 × 10−3 | 125 |
| Sc η | A | 0.96 | 3dMeA | 0.025 | 3.02 × 10−2 | 125 |
aHs, Homo sapien; Sc, S. cerevisiae.
bInsertion opposite 3dMeA relative to the insertion opposite an undamaged A.
cVmax/Km opposite 3dMeA relative to the Vmax/Km opposite 3dMeA by Hs polα.
dIn the presence of 0.25 mM MgCl2.
Very recently, we discovered that the catalytic activity of polι in vitro is dramatically enhanced in the presence of low concentrations of Mg2+ or Mn2+ (27). Indeed, polι-dependent incorporation opposite the 3dMeA lesion increased significantly when comparing primer extension in 0.25 mM versus 5 mM MgCl2 and lesion bypass was greatly stimulated in the presence of 0.25 mM MnCl2 (Figure 4). Similar to studies with other B-family polymerases (28), low levels of Mn2+ also appeared to stimulate human polδ's activity in the primer extension assays with the undamaged template, as well as enable a small amount of incorporation and extension beyond the 3dMeA lesion (Figure 4).
Figure 4.
Ability of human DNA polymerases δ and ι to bypass 3-deaza-3-methyl adenine in the presence of low Mg2+/Mn2+ in vitro. Standard reactions contained 100 μM all 4 dNTPs and lasted for 5 min at 37°C. Reactions contained 5 nM polδ and 4 nM polι. The nucleotide sequence of the template DNA is shown on the left-hand side of the gel. The ‘A’ in bold font is either undamaged (left-hand panel) or 3-deaza-3-methyl adenine (3dMeA; right-hand panel). Track 1, No dNTPs; Track 2, 0.25 mM MgCl2; Track 3, 5 mM MgCl2; Track 4, 0.25 mM MnCl2. As clearly seen, the 3dMeA lesion is a strong block to replication by human DNA polymerases δ even in the presence of Mn. In contrast, polι-dependent incorporation opposite the lesion is stimulated by 0.25 mM MgCl2 and significant bypass is observed in the presence of 0.25 mM MnCl2.
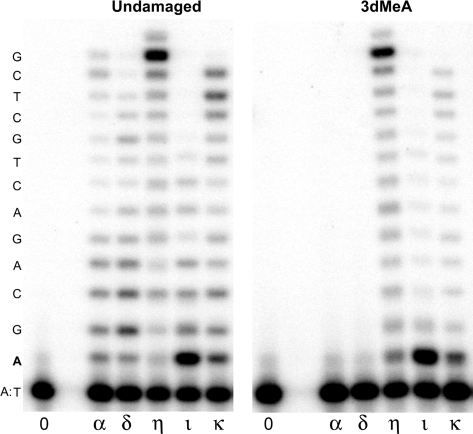
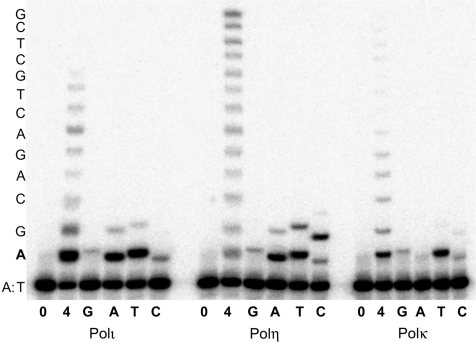
Next, we examined the single nucleotide insertion profile opposite the 3dMeA lesion promoted by polη, polι and polκ (Figure 5) and discovered that both polη and polι are error-prone, in that they readily misincorporate A opposite the 3dMeA lesion. Indeed, the ability of both polymerases to incorporate A opposite 3dMeA is consistent with the increase in A:T to T:A transversions observed in vivo in mice exposed to MMS (29). In contrast, polκ appears to be fairly accurate, as it primarily inserts T opposite 3dMeA. Analysis of the steady-state kinetics for each enzyme (Table 2) reflects the results shown in Figure 5. Polι and polη misinsert A opposite 3dMeA with a frequency of 0.46 and 0.48 relative to incorporation of the correct base T, respectively. In contrast, polκ is 10-fold more accurate than either polη or polι and misincorporates A opposite 3dMeA with a frequency of 0.04. Each polymerase appears to have higher than expected efficiency of inserting C, but this may simply occur as a consequence of the local sequence context, since the next 5′ template base is a G. While at first glance all three of the Y-family polymerases appear to be error-prone, they are, in fact, more accurate than polα, which actually misincorporates A opposite 3dMeA 4-fold better than T, in the steady-state assays (Table 2).
Figure 5.
Ability of human DNA polymerases η, ι and κ to (mis)incorporate opposite 3-deaza-3-methyl adenine. Standard reactions contained 100 μM all 4 dNTPs (4), or each nucleotide separately (G, A, T, C) and lasted for 5 min at 37°C. Reactions contained 4 nM polη, 6 nM polι and 4 nM polκ. The nucleotide sequence of the template DNA is shown on the left-hand side of the gel. The ‘A’ in bold font indicates the location of the 3dMeA lesion.
Table 2.
Fidelity of nucleotide insertion of opposite 3dMeA by various eukaryotic polymerases
| Polymerasea | Incoming nucleotide | Vmax/Km (μM−1 min−1) | finc |
|---|---|---|---|
| Hs α | A | 0.0008 | 4 |
| T | 0.0002 | 1 | |
| Hs η | G | 0.017 | 2.3 × 10−1 |
| A | 0.035 | 4.8 × 10−1 | |
| T | 0.073 | 1 | |
| C | 0.020 | 2.7 × 10−1 | |
| Hs ιb | G | 0.017 | 7.0 × 10−2 |
| A | 0.11 | 4.6 × 10−1 | |
| T | 0.24 | 1 | |
| C | 0.018 | 7.5 × 10−2 | |
| Hs κ | G | 0.002 | 9.9 × 10−2 |
| A | 0.001 | 4.4 × 10−2 | |
| T | 0.025 | 1 | |
| C | 0.003 | 1.3 × 10−1 | |
| Sc η | G | 0.002 | 6.2 × 10−2 |
| A | 0.004 | 1.6 × 10−1 | |
| T | 0.025 | 1 | |
| C | 0.005 | 2.2 × 10−1 |
aHs, Homo sapien; Sc, S. cerevisiae.
bIn the presence of 0.25 mM MgCl2.
Finally, we examined the ability of polη, polι and polκ to extend from a base paired with 3dMeA. The primer terminus was either a ‘correctly’ paired T:3dMeA, or was an A:3dMeA mispair (Table 3). Both polη and polκ extended the correctly paired T:3dMeA primer terminus relatively well and did so with an efficiency of ∼8–10% of that compared to a normal T:A base-pair (Table 3). In contrast, polι only extended the T:3dMeA primer with an efficiency of about 4% relative to an undamaged base-pair. Both polι and polκ extended the A:3dMeA mispair ∼3- to 4-fold less efficiently than the T:3dMeA base-pair. In contrast, human polη actually extended the A:3dMeA mispair slightly better than the correctly paired T:3dMeA (Table 3).
Saccharomyces cerevisiae polη is important in tolerating MMS-induced damage in the absence of MAG1
Based on our in vitro findings with the human Y-family polymerases, we were eager to determine if Y-family polymerases play a role in tolerating 3MeA in vivo. We chose to use S. cerevisiae as a model because it has a limited number of DNA polymerases compared with higher eukaryotes. Saccharomyces cerevisiae polη is encoded by the RAD30 gene, and it is reported that rad30Δ strains are somewhat sensitive to MMS (30,31). However, in the W303 background, a RAD30 disruption is not sensitive to MMS (Figure 6A). Mag1 is the only DNA glycosylase that repairs 3MeA in S. cerevisiae and disruption of the MAG1 gene makes yeast highly sensitive to methylating agents, such as MMS. In order to determine if S. cerevisiae polη is involved in tolerating lesions normally repaired by Mag1 (i.e. 3MeA), we generated a rad30Δ mag1Δ strain. Interestingly, the double mutant is more sensitive to MMS than the mag1Δ strain, suggesting that polη may help facilitate bypass of persisting 3MeA lesions in vivo (Figure 6A). Previous studies have shown that polζ is responsible for most MMS-induced mutagenesis (32,33), and deletion of REV3 (encoding the catalytic subunit of polζ) further sensitizes mag1Δ strains to MMS (Figure 6B). Therefore, it is possible that both polη and polζ are important for survival after treatment with MMS in a mag1Δ background (Figure 6B). Indeed, the triple mutant is significantly more sensitive than either double mutant (Figure 6B). This suggests that polζ and polη act in independent repair pathways to tolerate unrepaired base damage caused by MMS. Similar observations and conclusions were recently drawn by Johnson et al. (34).
Figure 6.
Survival of S. cerevisiae exposed to MMS. Exponentially growing strains of S. cerevisiae were exposed to 0.25% MMS for 10, 20 or 30 min, washed and subsequently plated on YPAD for 5 days at 30°C. (A) Disruption of REV3 makes S. cerevisiae mildly sensitive to MMS, but disruption of RAD30 has no observable effect on MMS sensitivity. (B) Disruption of MAG1 sensitizes S. cerevisiae to MMS, and disruption of RAD30 sensitizes the strain to MMS, indicating that the Rad30 encoded polη helps protect S. cerevisiae from lesions that are normally repaired by Mag1. (C) Disruption of REV3 also sensitizes a mag1Δ strain to MMS, and the disruption of both RAD30 and REV3 synergistically enhances the lethality of MMS, indicating that polη and polζ may operate in separate pathways to repair lesions normally removed from the genome by Mag1. Three independent isolates were tested for each strain and standard deviations, which were all below 1 log of survival have been omitted for clarity.
To determine which TLS polymerases in S. cerevisiae are capable of bypassing unrepaired 3MeA, we assayed the ability of S. cerevisiae polη, polζ, Rev1 and polζ in conjunction with Rev1 to bypass 3dMeA in vitro (Figure 7). Similar to human polη, S. cerevisiae polη bypasses the 3dMeA lesion reasonably efficiently. In contrast, polζ exhibits a much weaker ability to bypass the lesion (Figure 7, left-hand panel). Rev1, a dCMP transferase that is necessary for the function of polζ in vivo, also has minimal activity on either the undamaged A-template, or 3dMeA-containing template, as well as having little, to no stimulatory effect, on polζ's ability to bypass the 3dMeA lesion. Rev1 is clearly catalytically active under our assay conditions, as the enzyme is able to insert C opposite an undamaged template G, as well as further stimulate polζ activity (Figure 7, right-hand panel). Our in vitro data, combined with the MMS sensitivities of the mag1Δ rad30Δ and the mag1Δ rad30Δ rev3Δ strains therefore supports the idea that polη replicates past unrepaired 3MeA lesions in the absence of Mag1.
Figure 7.
Ability of S. cerevisiae DNA polymerases to bypass 3-deaza-3-methyl adenine in vitro. Standard reactions contained 100 μM all 4 dNTPs and lasted for 20 min at 30°C. Reactions contained 4 nM polη, 6 nM polζ and 15 nM Rev1. The nucleotide sequence of the DNA templates is shown on the left-hand side of each gel. Left-hand panels: the ‘A’ in bold font is either undamaged adenine or 3-deaza-3-methyl adenine. As can be seen, polη bypasses the 3dMeA lesion much more efficiently than polζ. Rev1 has negligible activity on either template, and did not appreciably stimulate the activity of polζ on the 3dMeA template. Right-hand panels: both Rev1 and polζ are catalytically active, as they are able to incorporate dCMP opposite undamaged G. Rev1 also stimulates polζ in the presence of dCTP alone as well as in the presence of all 4 dNTPs.
Human Y-family polymerases rescue the MMS sensitivity of mag1Δ rad30Δ rev3Δ strains of S. cerevisiae
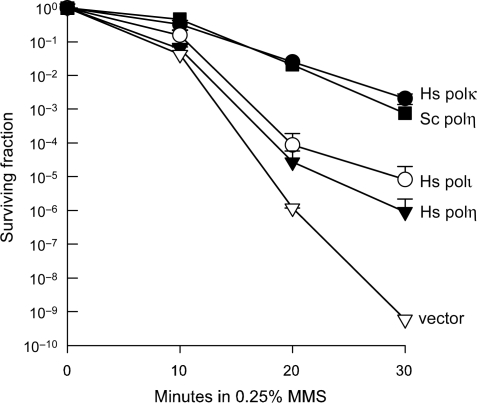
The enhanced MMS sensitivity of the mag1Δ rad30Δ rev3Δ strain gave us an opportunity to test the ability of human polη, polι and polκ to bypass alkylation damage in vivo. When each human polymerase (as well as S.cerevisiae polη, as a control), was expressed from a galactose inducible promoter in the triple mutant, we discovered that all of the human polymerases rescued the MMS-sensitivity of the mag1Δ rad30Δ rev3Δ strain, albeit to varying degrees (Figure 8). Quite remarkably, expression of human polκ confers MMS resistance on the mag1Δ rad30Δ rev3Δ strain to the same extent as overproducing S. cerevisiae polη. Both human polη and polι also confer MMS-resistance, but to a lesser degree than human polκ or S. cerevisiae polη.
Figure 8.
Human Y-family polymerases can restore MMS-resistance to a normally MMS-sensitive rad30Δ rev3Δ mag1Δ strain of S. cerevisiae. Exponentially growing strains of S. cerevisiae harboring plasmids expressing human polymerases η, ι and κ or S. cerevisiae polη under the control of a galactose inducible promoter were induced in complete synthetic galactose media without leucine. Media was removed, and cells were washed and exposed to 0.25% MMS for 10, 20 or 30 min, washed and subsequently plated on complete synthetic galactose plates lacking leucine for 5 days at 30°C. Three independent isolates were tested for each strain and standard deviations, which were all below 1 log of survival, have been omitted for clarity. As clearly seen, human polη and ι can restore MMS-resistance to the normally MMS-sensitive strain, but the greatest effect was observed with human polκ, which was as efficient as native S. cerevisiae polη in restoring MMS-resistance.
In some regard, it is really quite amazing that the human polymerases are able to confer MMS-resistance in the heterologous yeast survival assay, given the myriad of protein interactions that are believed to be required for the activity of the polymerases in vivo (35). Clearly, most of these protein–protein interactions must be conserved throughout evolution for the human polymerases to be able to function in S. cerevisiae. However, it is unlikely that these protein–protein interactions occur with the same efficiency in the heterologous system, and as a result, it is possible that the ability of human polι to restore MMS-resistance in S. cerevisiae is compromised by weakened protein–protein interactions with S. cerevisiae's TLS accessory proteins. The same cannot be said of human polη's inability to restore MMS-resistance to the same extent as S. cerevisiae polη, as human polη has previously been shown to fully complement the UV-sensitivity attributed to a polη-deficiency in a rad52Δ rad30Δ S. cerevisiae strain (36).
DISCUSSION
We have described a novel procedure for the synthesis of a phosphoramidite that is a stable 3-deaza analog of 3-methyl-2′-deoxyadenosine (Figure 1). By using this analog in replication assays, we provide the most direct evidence currently available that 3MeA is a significant block to two of the three main replicases in eukaryotes, namely polα and polδ. Furthermore, we demonstrate that three human Y-family polymerases (polη, polι and polκ) are capable of insertion opposite the 3dMeA lesion, as well as extension beyond the modified base (Figures 2–4), with polκ being the most accurate and polη the most efficient in vitro (Tables 1 and 2). Similarly, S. cerevisiae polη bypassed the 3dMeA lesion with the greatest efficiency of the Y-family polymerase assayed (Table 1), whilst polζ showed little ability to traverse the lesion in vitro (Figure 6A). Human polη, S. cerevisiae polη and human polκ, all extended bases incorporated opposite 3dMeA with an efficiency of 8–14% relative to an undamaged primer terminus (Table 3). Human polη did not discriminate between a correctly paired, or mispaired 3dMeA primer terminus, while human polκ and S. cerevisiae polη both preferred to extend the correctly paired T:3dMeA primer terminus 3- to 4-fold better then the A:3dMeA mispair. Polι extended the T:3dMeA base-pair poorly, but like human polκ and S. cerevisiae polη preferred the correctly paired primer terminus over the mispair.
Our kinetic data on the ability of human pols η, ι and κ to misinsert bases opposite a 3dMeA lesion in vitro does not agree well with a recent report in which 3MeA was modeled into the active site of the respective enzymes (34). Based upon molecular modeling it was hypothesized that polκ and polη should be able to insert a base opposite the 3MeA lesion equally as well as opposite an undamaged base. However, while significantly better than human polα, human polκ and polη inserted a base opposite 3dMeA with an efficiency of ∼0.2–2% of that opposite an undamaged base, suggesting some steric hindrance of the 3dMeA lesion in the active site of the respective Y-family enzymes. Similarly, it was also hypothesized that polι should be able to extend a T:3MeA base pair efficiently (34), but in our hands, this only occurred with an efficiency of about 4% of that of an undamaged base pair.
To examine the role of Y-family polymerases in tolerating 3MeA in vivo, we utilized strains of S. cerevisiae that carried a Rad30 (polη) deletion. At least in the wild-type W303 and CL1265-7C backgrounds (data for CL1265-7C not shown), the absence of polη did not appear to render the strain sensitive to MMS (Figure 6A). However, in the S288C background, a mild sensitivity has been previously reported (37). Since a large number of genes are known to be important for tolerating MMS in yeast (31), it is possible that subtle genetic differences between the W303 and the S288C backgrounds might account for the discrepancy between our observations and the data reported by others. Thus, despite the fact that S. cerevisiae polη bypasses the 3dMeA lesion in vitro (Figure 7), it does not appear to play a primary role in protecting wild-type cells from the cytotoxic effects of alkylation damage in vivo. Presumably such observations can be explained by the fact that 3MeA is not only intrinsically labile, but is efficiently removed from the genome by the Mag1 glycosylase (38). Interestingly, S. cerevisiae strains lacking both Mag1 and polη are significantly more sensitive to the cytotoxic effects of MMS than a wild-type strain (Figure 6B). We believe that such observations reveal an important role for polη in the bypass of persisting 3MeA lesions in vivo. It is also possible that the increased MMS-sensitivity may be partially due to a requirement for polη-dependent bypass of abasic sites. However, previous studies indicate that polη has limited ability to traverse an abasic site in vitro (39) and as a consequence, it is believed that polη plays only a minor role in the bypass of abasic site in vivo (40).
While polζ showed little ability to bypass the 3dMeA lesion in vitro, a rev3Δ strain nevertheless exhibited mild MMS-sensitivity in vivo (Figure 6A) (41). However, it should be noted that the strain is proficient for Mag1 and it is conceivable that the MMS sensitivity is actually due to an inability to bypass abasic lesions generated through the actions of the Mag1 glycoslyase, rather than defects in the bypass of 3MeA (32,33). The idea that polη and polζ potentially act in two separate pathways to facilitate bypass of 3MeA and abasic sites, respectively, is supported by the fact that the mag1Δ rad30Δ rev3Δ triple mutant is considerably more MMS-sensitive than either the mag1Δ rad30Δ or mag1Δ rev3Δ strains (Figure 6C).
The enhanced MMS sensitivity of the mag1Δ rad30Δ rev3Δ triple mutant allowed us to assay the role of human polη, polι and polκ in the tolerance of alkylation damage in vivo. While both expression of polη and polι increased MMS-resistance, expression of polκ resulted in MMS-resistance that was only rivaled by overexpression of endogenous S. cerevisiae polη (Figure 8). We believe that our data reflects how these enzymes might participate in the tolerance of alkylation damage in higher eukaryotes. Indeed, polκ appears to be important for survival after MMS exposure in both polκ-deficient MEFs and in polκ-deficient DT40 chicken lymphoblasts (42), and in both cases, it is assumed that the BER pathway in these cell lines remains fully functional. Furthermore, the role for polκ/polIV-like polymerases in tolerating cellular alkylation damage appears to be well conserved throughout evolution, as it has recently been reported that Escherichia coli dinB (polIV)-deficient strains are considerably more sensitive to MMS damage than wild-type strains (43).
The role of polη in tolerating MMS-induced lesions in vivo appears less clear. Polη appears to be important for budding yeast to tolerate MMS-induced damage, but only in the absence of BER (Figure 6A), and a similar situation may arise in human cells. Individuals with the variant form of Xeroderma Pigmentosum (XP-V) lack functional polη, and are susceptible to sunlight-induced skin cancer and while cells from these individuals are mildly sensitive to ultraviolet light, they are not sensitive to methylating agents, such as MMS (44).
The role of polι in the TLS of alkylation damage in mammals remains enigmatic. The Poli gene in the 129-derived inbred strain of mice has a stop codon in the second exon, effectively making the mice homozygous Poli(−/−) (45). Mice and embryonic stem cell lines derived from 129-derived strains are widely used in the study of DNA repair and mutagenesis, and appear to have no obvious sensitivity to methylating agents. However, it is possible that several ‘knockout’ mice generated using 129-derived embryonic stem cells could be ‘double knockouts’ for both Poli and the target gene of interest (46,47). Of direct importance to our current study, is the fact that two separate groups generated Aag(−/−) mice and cell lines from 129-derived embryonic stem cells (29,48). Interestingly, there were differences between the two studies in the sensitivity of the mice and MEFS to various alkylating agents. Elder et al., found that the Aag(−/−) primary fibroblasts exhibited mild sensitivity to MMS, but not bischroroethyl nitrosourea or mitomycin C, while Engelward and colleagues, who generated homozygous Aag(−/−) cells directly from a 129-derived embryonic stem line, observed that the MEFS were hypersensitive to MMS, bischloroethyl nitrosourea and mitomycin C. Essentially, the cells used by Elder et al. (29), were Aag(−/−) while those used by Engelward et al. (48), were likely to have been Aag(−/−), Poli(−/−). A subsequent study by Sobol and colleagues (49) found that cells independently derived from Aag(−/−) were not sensitive to MMS at all. Thus, subtle genotypic strain differences could readily account for the various phenotypes. As a consequence, it will be interesting to assay the MMS sensitivity of congenic C57Bl6-derived mice lacking Aag and Poli, to determine if polι plays a role in protecting mammalian cells from alkylation-induced DNA damage.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by funds from the NICHD/NIH Intramural Research Program. We thank Alexandra Vaisman for help preparing the figures and Haruo Ohmori for kindly providing the polκ plasmid, pHSE2. Funding to pay the Open Access publication charges for this article was provided by the NICHD/NIH Intramural Research Program.
Conflict of interest statement. None declared.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood R, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd. Washington, DC: ASM Press; 2006. [Google Scholar]
- 2.Hoffmann GR. Genetic effects of dimethyl sulfate, diethyl sulfate, and related compounds. Mutat. Res. 1980;75:63–129. doi: 10.1016/0165-1110(80)90028-7. [DOI] [PubMed] [Google Scholar]
- 3.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat. Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 4.Monti P, Iannone R, Campomenosi P, Ciribilli Y, Varadarajan S, Shah D, Menichini P, Gold B, Fronza G. Nucleotide excision repair defect influences lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent in the absence of base excision repair. Biochemistry. 2004;43:5592–5599. doi: 10.1021/bi035968x. [DOI] [PubMed] [Google Scholar]
- 5.Memisoglu A, Samson L. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 2000;182:2104–2112. doi: 10.1128/jb.182.8.2104-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao W, Chow BL, Rathgeber L. The repair of DNA methylation damage in Saccharomyces cerevisiae. Curr. Genet. 1996;30:461–468. doi: 10.1007/s002940050157. [DOI] [PubMed] [Google Scholar]
- 7.Plosky B, Samson L, Engelward BP, Gold B, Schlaen B, Millas T, Magnotti M, Schor J, Scicchitano DA. Base excision repair and nucleotide excision repair contribute to the removal of N-methylpurines from active genes. DNA Repair. 2002;1:683–696. doi: 10.1016/s1568-7864(02)00075-7. [DOI] [PubMed] [Google Scholar]
- 8.Engelward BP, Allan JM, Dreslin AJ, Kelly JD, Wu MM, Gold B, Samson LD. A chemical and genetic approach together define the biological consequences of 3-methyladenine lesions in the mammalian genome. J. Biol. Chem. 1998;273:5412–5418. doi: 10.1074/jbc.273.9.5412. [DOI] [PubMed] [Google Scholar]
- 9.Singer B, Brent TP. Human lymphoblasts contain DNA glycosylase activity excising N-3 and N-7 methyl and ethyl purines but not O6-alkylguanines or 1-alkyladenines. Proc. Natl Acad. Sci. USA. 1981;78:856–860. doi: 10.1073/pnas.78.2.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison A, Christensen RB, Alley J, Beck AK, Bernstine EG, Lemontt JF, Lawrence CW. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J. Bacteriol. 1989;171:5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nelson JR, Lawrence CW, Hinkle DC. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 12.Ohmori H, Friedberg EC, Fuchs RPP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 13.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 14.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase δ using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 15.Tissier A, McDonald JP, Frank EG, Woodgate R. polι, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 16.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, Iwai S, Hanaoka F. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaisman A, Takasawa K, Iwai S, Woodgate R. DNA polymerase ι-dependent translesion replication of uracil containing cyclobutane pyrimidine dimers. DNA Repair. 2006;5:210–218. doi: 10.1016/j.dnarep.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 18.Creighton S, Bloom LB, Goodman MF. Gel fidelity assay measuring nucleotide misinsertion, exonucleolytic proofreading, and lesion bypass efficiencies. Methods Enzymol. 1995;262:232–256. doi: 10.1016/0076-6879(95)62021-4. [DOI] [PubMed] [Google Scholar]
- 19.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 20.McDonald JP, Levine AS, Woodgate R. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics. 1997;147:1557–1568. doi: 10.1093/genetics/147.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald JP, Rapic-Otrin V, Epstein JA, Broughton BC, Wang X, Lehmann AR, Wolgemuth DJ, Woodgate R. Novel human and mouse homologs of Saccharomyces cerevisiae DNA polymerase η. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- 22.Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer B. N-nitroso alkylating agents: formation and persistence of alkyl derivatives in mammalian nucleic acids as contributing factors in carcinogenesis. J. Natl Cancer Inst. 1979;62:1329–1339. [PubMed] [Google Scholar]
- 24.Chakravarti D, Ibeanu GC, Tano K, Mitra S. Cloning and expression in Escherichia coli of a human cDNA encoding the DNA repair protein N-methylpurine-DNA glycosylase. J. Biol. Chem. 1991;266:15710–15715. [PubMed] [Google Scholar]
- 25.Saparbaev M, Kleibl K, Laval J. Escherichia coli, Saccharomyces cerevisiae, rat and human 3-methyladenine DNA glycosylases repair 1,N6-ethenoadenine when present in DNA. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc. Natl Acad. Sci. USA. 2000;97:13573–13578. doi: 10.1073/pnas.97.25.13573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank EG, Woodgate R. Increased catalytic activity and altered fidelity of DNA polymerase ι in the presence of manganese. J. Biol. Chem. 2007;282:24689–24696. doi: 10.1074/jbc.M702159200. [DOI] [PubMed] [Google Scholar]
- 28.Villani G, Tanguy Le Gac N, Wasungu L, Burnouf D, Fuchs RP, Boehmer PE. Effect of manganese on in vitro replication of damaged DNA catalyzed by the herpes simplex virus type-1 DNA polymerase. Nucleic Acids Res. 2002;30:3323–3332. doi: 10.1093/nar/gkf463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elder RH, Jansen JG, Weeks RJ, Willington MA, Deans B, Watson AJ, Mynett KJ, Bailey JA, Cooper DP, Rafferty JA, et al. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol. Cell Biol. 1998;18:5828–5837. doi: 10.1128/mcb.18.10.5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roush AA, Suarez M, Friedberg EC, Radman M, Siede W. Deletion of the Saccharomyces cerevisiae gene RAD30 encoding an Escherichia coli DinB homolog confers UV radiation sensitivity and altered mutability. Mol. Gen. Genet. 1998;257:686–692. doi: 10.1007/s004380050698. [DOI] [PubMed] [Google Scholar]
- 31.Begley TJ, Rosenbach AS, Ideker T, Samson LD. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 2002;1:103–112. [PubMed] [Google Scholar]
- 32.Johnson RE, Torres-Ramos CA, Izumi T, Mitra S, Prakash S, Prakash L. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 1998;12:3137–3143. doi: 10.1101/gad.12.19.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao W, Chow BL, Hanna M, Doetsch PW. Deletion of the MAG1 DNA glycosylase gene suppresses alkylation-induced killing and mutagenesis in yeast cells lacking AP endonucleases. Mutat. Res. 2001;487:137–147. doi: 10.1016/s0921-8777(01)00113-6. [DOI] [PubMed] [Google Scholar]
- 34.Johnson RE, Yu SL, Prakash S, Prakash L. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol. Cell. Biol. 2007;27:7198–7205. doi: 10.1128/MCB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl Acad. Sci. USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glick E, Vigna KL, Loeb LA. Mutations in human DNA polymeraseη motif II alter bypass of DNA lesions. EMBO J. 2001;20:7303–7312. doi: 10.1093/emboj/20.24.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao B, Xie Z, Shen H, Wang Z. Role of DNA polymerase η in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 2004;32:3984–3994. doi: 10.1093/nar/gkh710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berdal KG, Bjoras M, Bjelland S, Seeberg E. Cloning and expression in Escherichia coli of a gene for an alkylbase DNA glycosylase from Saccharomyces cerevisiae; a homologue to the bacterial alkA gene. EMBO J. 1990;9:4563–4568. doi: 10.1002/j.1460-2075.1990.tb07909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haracska L, Washington MT, Prakash S, Prakash L. Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem. 2001;276:6861–6866. doi: 10.1074/jbc.M008021200. [DOI] [PubMed] [Google Scholar]
- 40.Gibbs PEM, McDonald JP, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol η, Pol ζ, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct, and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–582. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Broomfield S, Chow BL, Xiao W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl Acad. Sci. USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takenaka K, Ogi T, Okada T, Sonoda E, Guo C, Friedberg EC, Takeda S. Involvement of vertebrate Polκ in translesion DNA synthesis across DNA monoalkylation damage. J. Biol. Chem. 2006;281:2000–2004. doi: 10.1074/jbc.M506153200. [DOI] [PubMed] [Google Scholar]
- 43.Bjedov I, Nag Dasgupta C, Slade D, Le Blastier S, Selva M, Matic I. Involvement of Escherichia coli DNA polymerase IV in tolerance of cytotoxic alkylating DNA lesions in vivo. Genetics. 2007;176:1431–1440. doi: 10.1534/genetics.107.072405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teo IA, Arlett CF. The response of a variety of human fibroblast cell strains to the lethal effects of alkylating agents. Carcinogenesis. 1982;3:33–37. doi: 10.1093/carcin/3.1.33. [DOI] [PubMed] [Google Scholar]
- 45.McDonald JP, Frank EG, Plosky BS, Rogozin IB, Masutani C, Hanaoka F, Woodgate R, Gearhart PJ. Identification of a nonsense mutation in DNA polymerase ι from 129-derived strains of mice and its effect on somatic hypermutation. J. Exp. Med. 2003;198:635–643. doi: 10.1084/jem.20030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald JP, Woodgate R. Letter to the editor. DNA Repair. 2003;2:1159–1160. [Google Scholar]
- 47.Sobol RW. DNA polymerase β null mouse embryonic fibroblasts harbor a homozygous null mutation in DNA polymerase ι. DNA Repair. 2007;6:3–7. doi: 10.1016/j.dnarep.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 49.Sobol RW, Kartalou M, Almeida KH, Joyce DF, Engelward BP, Horton JK, Prasad R, Samson LD, Wilson SH. Base excision repair intermediates induce p53-independent cytotoxic and genotoxic responses. J. Biol. Chem. 2003;278:39951–39959. doi: 10.1074/jbc.M306592200. [DOI] [PubMed] [Google Scholar]