Abstract
A nuclear magnetic resonance (NMR) investigation of a fragment of the nonreceptor Tec family tyrosine kinase Btk has revealed an intricate set of coupled monomer-dimer equilibria. The Btk fragment studied contains two consecutive proline-rich motifs followed by a single Src homology 3 (SH3) domain. We provide evidence for an asymmetric homodimer in which the amino-terminal proline sequence of one monomer contacts the opposite SH3 binding pocket, whereas the carboxy-terminal proline sequence of the other monomer is engaged by the second SH3 domain across the dimer interface. We show that the asymmetric homodimer structure is mimicked by a heterodimer formed in an equimolar mixture of complimentary mutants: one carrying mutations in the amino-terminal proline stretch; the other, in the carboxy-terminal proline motif. Moreover, a monomeric species characterized by an intramolecular complex between the amino-terminal proline motif and the SH3 domain predominates at low concentration. Association constants were determined for each of the competing equilibria by NMR titration. The similarity of the determined Ka values reveals a delicate balance between the alternative conformational states available to Btk. Thus, changes in the local concentration of Btk itself, or co-localization with exogenous signaling molecules that have high affinity for either proline sequence or the SH3 domain, can significantly alter species composition and regulate Btk kinase activity.
Keywords: Bruton's tyrosine kinase (Btk), SH3, NMR, asymmetric homodimer, intramolecular, self-association
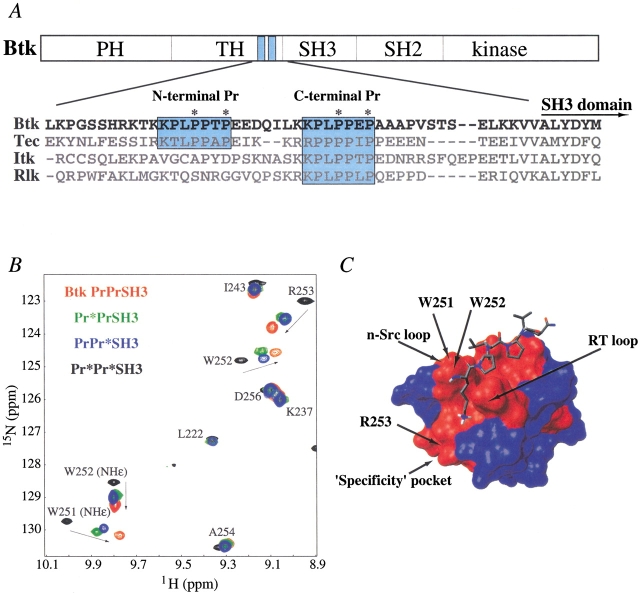
Phosphorylation of substrates by the catalytic action of protein kinases is a key chemical modification responsible for cellular signal transduction. Unlike the well-studied Src family of kinases, issues of structure and function are only beginning to emerge for the immunological kinases of the Tec family (Rawlings and Witte 1995; Mano 1999). To understand the mechanism of Tec kinase catalytic regulation, we have conducted a structural analysis of Bruton's tyrosine kinase (Btk), the protein for which genetic defects cause the human disease X-linked agammaglobulinemia (XLA; Tsukada et al. 1993; Duriez et al. 1994; Ohta et al. 1994). The domain structure of Btk (Vetrie et al. 1993) is shown in Figure 1A ▶. Typical of the Tec kinases, Btk contains a proline-rich region adjacent to a conserved Src homology 3 (SH3) domain. SH3 domains are ubiquitous, noncatalytic adaptor modules that recognize and bind to proline-rich sequences (for review, see Kuriyan and Cowburn 1997). Structural data have recently clarified the role of SH3 domains in the regulation of Src family kinases (Sicheri et al. 1997; Xu et al. 1997). Intramolecular binding events involving both the SH3 and the Src homology 2 (SH2) domains of Src kinases alter the conformation of active site residues within the kinase domain to down-regulate kinase activity. The Tec kinases, however, lack the carboxy-terminal regulatory phosphorylation site of the Src kinases (the intramolecular target for the Src SH2 domain) and instead contain an extended amino-terminal region consisting of a Pleckstrin homology (PH) domain (Lemmon et al. 1996) and a Tec homology (TH) domain (Vihinen et al. 1994). The differences between domain structures of the Src and Tec kinase families indicate that control of Tec kinase activity is most likely accomplished via a distinct mechanism.
Fig. 1.
(A) Domain structure of Btk and sequence alignment of a portion of Btk (residues 175 to 226) and related Tec family members (Bolen 1995) Tec, Itk, and Rlk (accession nos. XM010110, P24604, A43030, and A55631, respectively). The amino- and carboxy-terminal proline-rich (Pr) sequences within the TH domain are delimited by blue boxes. (Itk and Rlk each contain only a single Pr region.) Asterisks indicate the specific proline residues that are mutated to alanine in the Pr* mutants. Thus, Pr*Pr*SH3 is used to denote a double mutant in which the two critical prolines in both motifs have been mutated to alanine. Likewise, the single mutants, Pr*PrSH3 and PrPr*SH3, each contain two Pro-to-Ala substitutions in the amino- and carboxy-terminal Pr regions, respectively. The amino-terminal domain boundary of the SH3 domain is indicated in the alignment. (B) Overlay of a portion of four 1H-15N heteronuclear single quantum coherence (HSQC) spectra of recombinant PrPrSH3 and corresponding mutants. Cross-peaks for wild-type PrPrSH3 protein are red, those for the amino-terminal proline mutant (Pr*PrSH3) are green, those for the carboxy-terminal mutant (PrPr*SH3) are blue, and those for the construct in which both proline motifs are altered (Pr*Pr*SH3) are black. The concentration of each sample is 1 mM. Spectral assignments for the Btk SH3 domain have been reported previously (Hansson et al. 1998). Residues that define the SH3 binding pocket show extensive chemical shift perturbations (denoted with arrows) when the indicated proline residues are mutated to alanine. Residues outside of the SH3 binding pocket appear at the same chemical shift regardless of mutations. (C) Proline-rich peptide bound to the Btk SH3 domain (Tzeng et al. 2000). Residues showing significant chemical shift differences (>0.15 ppm, indicated in red) between monomeric Pr*Pr*SH3 and wild-type PrPrSH3 define the canonical SH3 binding groove (Yu et al. 1992). The n-Src and RT loops lining the proline binding groove are labeled, as well as the specificity pocket that has been shown previously to contact ligand residues amino-terminal to the core proline motif (Feng et al. 1995). A similar analysis for each of the single mutants (Pr*PrSH3 and PrPr*SH3) reveals the same binding surface (not shown). Surface plot rendered using MolMol (Molecular Analysis and Molecular Display; Koradi et al. 1996) from the PDB file 1QLY.
Inspection of the primary amino acid sequences of the Tec family kinases (Fig. 1A ▶) reveals potentially important differences within the PH and TH domains. In particular, Btk contains a stretch of two class I proline-rich motifs (Feng et al. 1994) that conform to the consensus sequence of an SH3 ligand. It has been recently shown that a Btk fragment that contains the two proline regions and the SH3 domain (denoted here as PrPrSH3) self-associates in an intermolecular fashion (Hansson et al. 2001). This is in contrast to the T-cell analog Itk (Siliciano et al. 1992; Heyeck and Berg 1993), which contains a single proline motif and adopts a fold in which the proline region contacts the SH3 binding pocket in an intramolecular fashion (Andreotti et al. 1997). We now report on nuclear magnetic resonance (NMR) structural investigations of the Btk PrPrSH3 fragment that reveal the molecular details of self-association. We show that both intra- and intermolecular association modes are present for the Btk PrPrSH3 fragment, which raises interesting possibilities regarding the regulation of the Tec kinases.
Results
NMR analysis of Btk PrPrSH3 and corresponding mutants
1H-15N heteronuclear single quantum coherence (HSQC) spectra consist of cross-peaks at specific 1H and 15N chemical shifts that correspond to the amide groups of the protein under study. The position of each cross-peak is extremely sensitive to the chemical environment of the corresponding NH group and therefore reflects changes that accompany binding. A superposition of HSQC spectra (Mori et al. 1995) of the recombinant Btk PrPrSH3 fragment and the corresponding fragment in which the critical prolines in both proline stretches are mutated to alanine (Pr*Pr*SH3) is shown in Figure 1B ▶. It is well established that mutation of these proline residues abolishes SH3 affinity for the resulting amino acid sequence (Feng et al. 1994). In fact, the cross-peaks in the HSQC spectrum of a fragment consisting of the Btk SH3 domain alone are an exact subset of those of the double mutant Pr*Pr*SH3, indicating that the SH3 domain ligand binding pocket is unoccupied in this mutant (data not shown). The residues defined by the cross-peaks showing chemical shift differences between the double mutant and wild-type PrPrSH3 map out the contiguous, previously well-defined proline ligand binding pocket (Yu et al. 1992) of the Btk SH3 domain (Fig. 1C ▶).
To determine the precise role of the two proline sequences in mediating dimerization of PrPrSH3, we characterized two additional Btk `single' mutants, Pr*PrSH3 and PrPr*SH3 (see Fig. 1A ▶ caption), by NMR spectroscopy. First, NMR line widths were analyzed at high (1.5 to 2.0 mM) and low (0.03 to 0.05 mM) concentrations to determine the predominant aggregation state of the single mutants. At high concentration, average NMR line widths ranged from 24 to 31 Hz for both single mutants and wild-type PrPrSH3, whereas they ranged from 15 to 19 Hz for all three proteins at low concentrations. Line widths for Pr*Pr*SH3 were constant between 0.4 and 3.5 mM and ranged from 15 to 19 Hz. Thus, Btk fragments that contain at least one intact proline motif dimerize in solution, whereas the double mutant does not. To ascertain whether dimerization of the single mutants occurs via interactions mediated by the SH3 binding pocket contacting the proline-rich ligand, the chemical shift differences between each of the single mutants and Pr*Pr*SH3 were analyzed. Significant chemical shift perturbations occur for the same residues as were identified for wild-type Btk PrPrSH3 when compared to double mutant and therefore map out the same well-defined binding pocket of the SH3 domain (Fig. 1C ▶). However, the magnitude of the chemical shift changes for the single mutants compared with Pr*Pr*SH3 appears to be less than that for wild-type protein (Fig. 1B ▶).
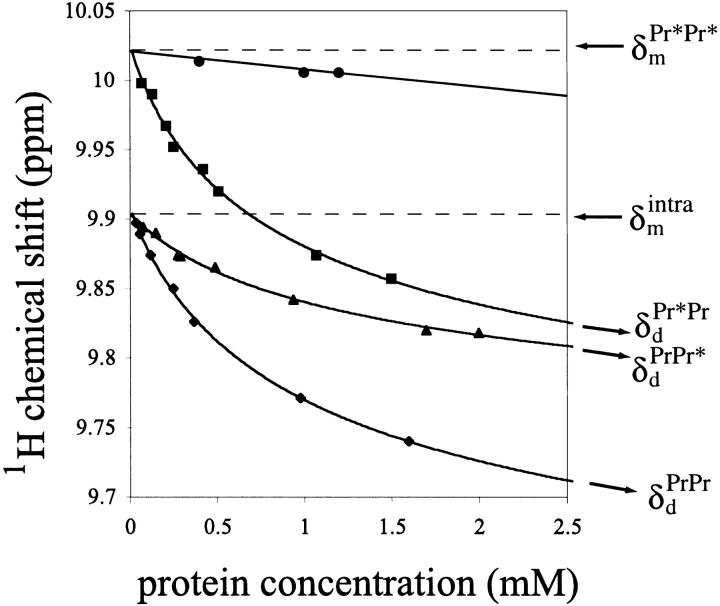
Progressive changes in the positions of 21 of 67 cross-peaks in the HSQC spectra of each of the proteins—PrPrSH3, Pr*PrSH3, and PrPr*SH3—were observed as protein concentration was varied. Representative plots of the proton resonance frequency of W251(NHɛ) versus protein concentration are shown in Figure 2 ▶ for the two single mutants, wild-type Btk PrPrSH3, and the monomeric `double' mutant Pr*Pr*SH3. For a system in fast exchange, the observed chemical shift is the weighted average of the chemical shifts of reporters in the two exchanging species, as given by Equation 1 (Cheng and Shirts 1985):
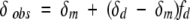 |
(1) |
where δobs, δm, and δd are the observed, monomeric (at infinite dilution) and dimeric (at infinite concentration) chemical shifts, respectively. The mole fraction of dimer (fd) is calculated according to a monomer-dimer equilibrium model (Equation 2) and is given by Equation 3 (Cheng and Shirts 1985)
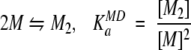 |
(2) |
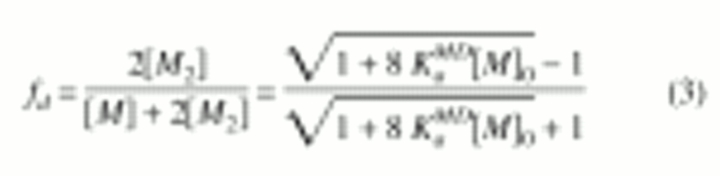 |
(3) |
where KaMD is the observed association constant (the MD superscript indicates that it was obtained based on a monomer-dimer equilibrium model), and the zero subscript indicates total protein concentration.
Fig. 2.
Concentration dependence of chemical shift for W251(NHɛ) in the context of wild-type PrPrSH3 (♦), Pr*PrSH3 (▪), PrPr*SH3 (▴), and Pr*Pr*SH3 (•) are shown. Data were fit to a monomer-dimer equilibrium model (Equations 2, 3). Extrapolated values of the chemical shift at infinite dilution for PrPr*SH3, and PrPrSH3(δmintra) as well as Pr*PrSH3 and Pr*Pr*SH3 (δmPr*Pr*) are shown. Extrapolated values of the chemical shift at infinite concentration for both single mutants (δdPrPr* and δdPr*Pr) and wild-type PrPrSH3 (δdPrPr) are also indicated. Very small chemical shift changes are observed for Pr*Pr*SH3, indicating nonspecific aggregation at high concentrations.
Observed association constants, δm, and δd values were determined by three parameter least-squares fitting of Equation 1 to the concentration dependence of the proton chemical shift for five different SH3 domain residues: W251(NHɛ; shown in Fig. 2 ▶), I264, N267, W252, and R253. The values of KaMD were then averaged for each protein, giving values of 0.5±0.1, 0.8±0.1, and 0.3±0.1 (mM−1) for wild-type PrPrSH3, Pr*PrSH3, and PrPr*SH3, respectively. For PrPrSH3, tryptophan fluorescence intensity (340 nm) was linear with respect to concentration over the range of 20 to 70 μM (data not shown). Therefore, negligible association occurs at these concentrations, in agreement with the association constants determined by NMR spectroscopy.
Analysis of the extrapolated monomeric chemical shifts (δm)
The system is in fast exchange, relative to the NMR time scale, and so the magnitude of the chemical shift perturbations for residues in the SH3 binding pocket can be used as a measure of relative SH3 occupancy by a proline ligand. Interestingly, the asymptotic limits of the binding curves (and therefore SH3 occupancy in the monomeric and dimeric forms) for each of the related Btk fragments are different. As can be seen in Figure 2 ▶, the monomeric chemical shifts (denoted δmintra) for PrPrSH3 and PrPr*SH3 are nearly equal. The corresponding values for the Pr*PrSH3 mutant and the double mutant (denoted δmPr*Pr*) are also nearly equal and are different from δmintra. This is the case for all SH3 residue frequencies that show significant concentration-dependent chemical shift changes.
To explain these results, we propose a structural model for Btk in which the amino-terminal proline sequence contacts the adjacent SH3 domain binding pocket in an intramolecular fashion in both wild-type PrPrSH3 and PrPr*SH3 (Fig. 3A ▶). The Btk intramolecular interaction competes directly with dimer formation because both binding events require occupancy of the same SH3 binding pocket. The value of δm for the Pr*PrSH3 mutant indicates that the carboxy-terminal proline stretch within Btk does not contact the SH3 pocket in an intramolecular fashion. The intramolecular association mode observed here for Btk is analogous to that observed previously for the homologous proline SH3-containing Itk fragment (Andreotti et al. 1997). However, the Itk fragment does not dimerize at higher concentrations.
Fig. 3.
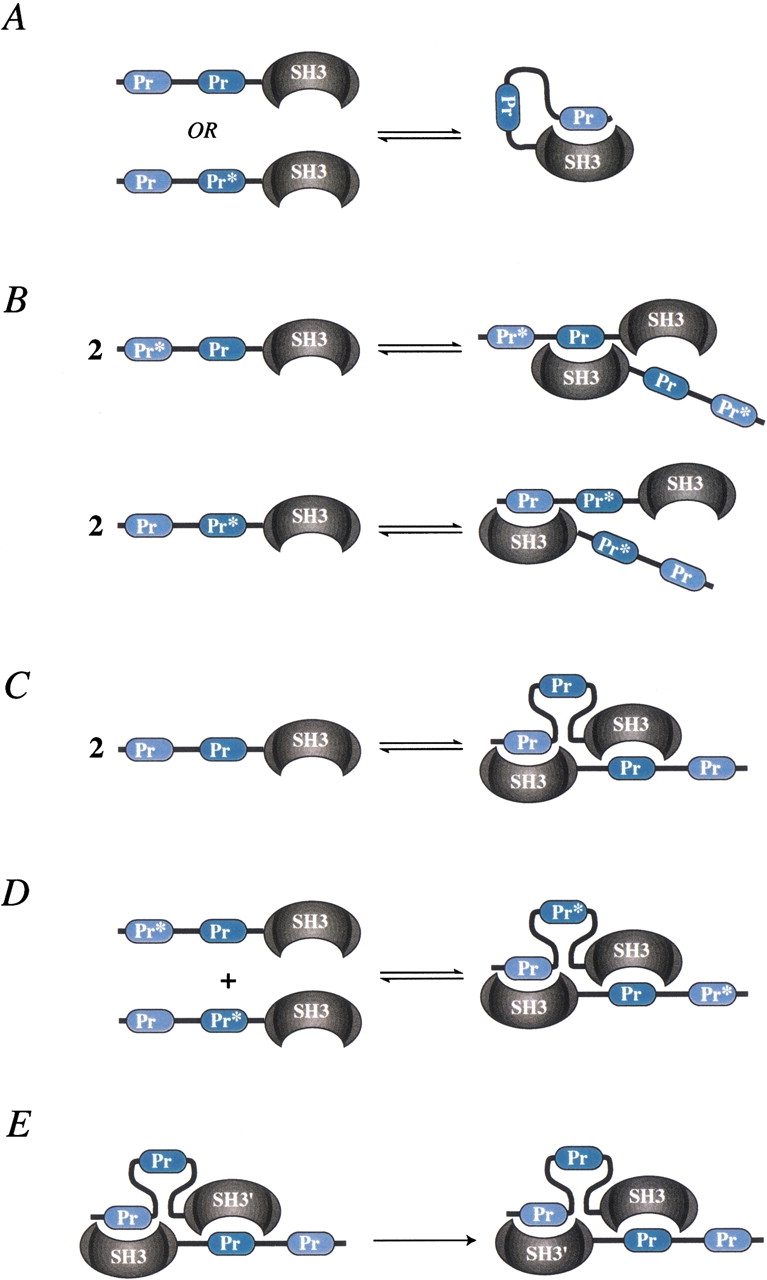
Schematic depictions of the PrPrSH3 fragments studied and the corresponding equilibria as determined by nuclear magnetic resonance. (A) The amino-terminal proline motif can occupy the SH3 binding groove in an intramolecular fashion. (B) Dimerization of each of the single mutants (Pr*PrSH3 and PrPr*SH3) results in partial SH3 occupancy. (C) Proposed asymmetric association mode of wild-type PrPrSH3. (D) Mixture of complimentary single mutants that mimic wild-type asymmetric dimer. (E) A symmetry operation on the proposed homodimer that interconverts the two subunits.
Analysis of the extrapolated dimeric chemical shifts (δd)
The extrapolated chemical shift values corresponding to infinite concentration (δd) also reveal differences among the Btk constructs studied. The limits of the chemical shift at high concentration for both single mutants (denoted δdPr*Pr and δdPrPr* in Fig. 2 ▶) are approximately the same, whereas the extent of chemical shift perturbation resulting from wild-type dimer formation (δdPrPr in Fig. 2 ▶) is significantly greater. This indicates that the population of occupied SH3 binding pockets of the single mutants is less (approximately half) than that of wild type in the dimer form. Proposed association modes for the single mutants are depicted in Figure 3B ▶ for Pr*PrSH3 and PrPr*SH3.
We have established that both Btk single mutants (Pr*PrSH3 and PrPr*SH3) and wild-type PrPrSH3 dimerize and that the extent of SH3 occupancy in the single mutants and wild type is different. Furthermore, the molecular surface mapped by chemical shift perturbations is the same in all three cases and corresponds to the SH3 proline binding pocket. If only a single proline stretch were responsible for the intermolecular association in wild-type PrPrSH3, then either the Pr*PrSH3 or the PrPr*SH3 mutant would not dimerize, whereas the other single mutant would behave similarly to wild type. Thus, the most plausible structural model for Btk PrPrSH3 dimerization is an asymmetric homodimer in which one of the two SH3 binding pockets in the dimer contacts the amino-terminal proline sequence, whereas the other SH3 domain binds to the carboxy-terminal proline sequence (Fig. 3C ▶).
Mixture of complimentary mutants mimic wild-type Btk PrPrSH3
To ascertain whether the PrPrSH3 fragment forms the proposed asymmetric homodimer, we performed NMR titration experiments using mixtures of complimentary single mutants. As we have already shown, the single mutants alone form a homodimer in which one of the SH3 proline binding sites is unoccupied (Fig. 3B ▶). However, for a mixture of single mutants (Pr*PrSH3 and PrPr*SH3), formation of the asymmetric heterodimer in a manner that mimics the putative wild-type asymmetric homodimer (i.e., having two occupied proline binding sites) is possible (Fig. 3D ▶).
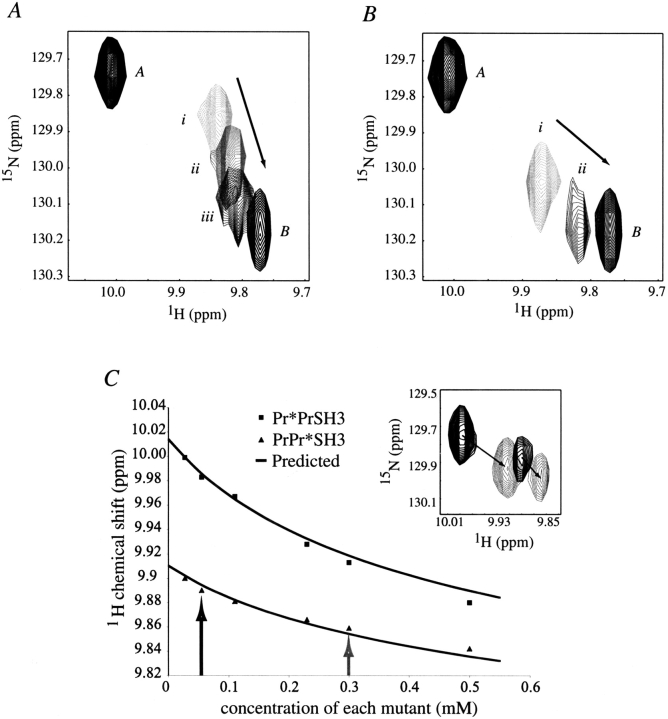
Addition of unlabeled single mutant (Pr*PrSH3) into a sample of the 15N-labeled complimentary mutant (PrPr*SH3) should shift the equilibrium toward occupied SH3 domain if asymmetric homodimerization occurs. Indeed, the position of the cross-peak corresponding to W251(NHɛ; Fig. 4A ▶) shifts significantly in the presence of 0.2 and then 0.4 equivalents of complimentary unlabeled Pr*PrSH3 mutant. Similar shifts are observed for all SH3 cross-peaks, corresponding to residues in the proline binding pocket. The direction of the observed chemical shift perturbation is toward that of occupied SH3 domain (i.e., that of 1 mM wild-type PrPrSH3). The only proline motif that can cause the observed shift is the carboxy-terminal stretch of the unlabeled mutant (Pr*PrSH3), which binds to the SH3 pocket of 15N-labeled PrPr*SH3 in an intermolecular fashion. In the reverse titration (addition of unlabeled PrPr*SH3 into a sample of 15N-labeled Pr*PrSH3), similar chemical shift perturbations are observed (Fig. 4B ▶). In this case, the amino-terminal proline stretch must be responsible for the observed shifts in the labeled SH3 domain. Combined, these data show that asymmetric dimer formation is possible.
Fig. 4.
(A) Overlay of five separately acquired heteronuclear single quantum coherence (HSQC) spectra showing the cross-peak for W251(NHɛ): 1.2 mM Pr*Pr*SH3 (A), 1 mM PrPr*SH3 (i), 1 mM PrPr*SH3 plus addition of 0.2 mM unlabeled Pr*PrSH3 (ii), 1 mM PrPr*SH3 plus a total of 0.4 mM unlabeled Pr*PrSH3 (iii), and 1 mM wild-type PrPrSH3 (B). Peaks A and B indicate the position of W251(NHɛ) for unbound SH3 domain and occupied SH3 domain, respectively. (B) A titration similar to A, but with a reversed labeling scheme: 1.2 mM Pr*Pr*SH3 (A), 1 mM Pr*PrSH3 (i), 1 mM Pr*PrSH3 plus addition of 1 mM unlabled PrPr*SH3 (ii), and 1 mM PrPrSH3 (B). Observed shifts are caused by protein-protein interaction and not by changes in salt concentration that result from addition of lyophilized protein sample to the NMR tube. (C) Data obtained from six separate HSQC experiments performed on equimolar mixtures of complimentary mutants (Pr*PrSH3 and PrPr*SH3). The concentration dependence for the proton chemical shift of W251(NHɛ) is shown, for cross-peaks corresponding to Pr*PrSH3 (▪) and PrPr*SH3 (▴). A pair of peaks is observed because of the two distinct species in solution. The predicted concentration behavior, resulting from an analysis of the four competing equilibria defined in Equations 5 through 8 is shown as a line and agrees well with the observed values. (Inset) An overlay of the W251(NHɛ) region of two HSQC spectra (mixtures of 0.3 mM of each mutant and 0.06 mM of each mutant; gray and black, respectively), shows two distinct cross-peaks as described. Points which represent the data shown in the inset are indicated by appropriately shaded arrows.
An additional experiment was conducted in which a 1 mM solution of 15N-labeled PrPr*SH3 was combined with an equivalent volume of 1 mM 15N-labeled Pr*PrSH3. This sample was then serially diluted, and HSQC spectra were collected. Two HSQC cross-peaks appear for each residue in the SH3 proline binding pocket (Fig. 4C ▶, inset), corresponding to the two proteins Pr*PrSH3 and PrPr*SH3. The appearance of two peaks for each residue is owing to the fact that the two species (Pr*PrSH3 and PrPr*SH3) are not in chemical exchange and have different resonance frequencies at low concentration (see Fig. 2 ▶). PrPr*SH3 can associate in an intramolecular fashion, whereas Pr*PrSH3 can not, giving rise to distinct values. As protein concentration is increased, the resonance frequency difference between the two peaks diminishes, and both cross-peaks shift toward the frequency of occupied SH3. However, the concentration-dependent behavior of the cross-peaks corresponding to each protein in the mixture differs from that observed when the single mutants are analyzed separately (as in Fig. 2 ▶). The difference arises from the fact that Pr*PrSH3 and PrPr*SH3 can interact with each other and form the putative asymmetric heterodimer. Given the self-association constants for the single mutants and wild-type PrPrSH3, the behavior of the mixture can be predicted.
Quantitative analysis of the competing equilibria
The predicted concentration dependence of the resonance frequencies for each cross-peak and the observed experimental values are plotted in Figure 4C ▶. The equilibrium model used to predict the concentration dependence of the chemical shifts for the mixture of single mutants was developed by first determining K`a, the intramolecular association constant of either PrPr*SH3 or PrPrSH3. Because both SH3 pockets are occupied in wild-type PrPrSH3 dimer, δdPrPr (see Fig. 2 ▶) is the chemical shift of a species with fully occupied SH3 pockets, whereas δPr*Pr*m represents the chemical shift of the unoccupied species. We can therefore calculate K`a as the ratio of occupied to unoccupied SH3 pockets in the monomer, as defined in Equation 4,
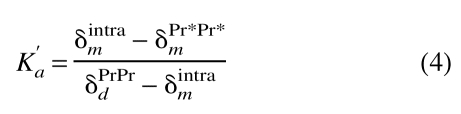 |
(4) |
where δPr*Pr*m is the chemical shift of Pr*Pr*SH3, and δPrPrd is the extrapolated chemical shift of dimeric PrPrSH3. δintram is the extrapolated chemical shift corresponding to either monomeric wild-type PrPrSH3 or monomeric PrPr*SH3. The determined values of K`a are 0.28±0.05 and 0.29±0.06 for Btk PrPrSH3 and PrPr*SH3, respectively.
We assume that the system is at equilibrium and that six species are present: PrPr*SH3 monomer intramolecularly bound, PrPr*SH3 monomer not intramolecularly bound, Pr*PrSH3 monomer, PrPr*SH3 homodimer, Pr*PrSH3 homodimer, and Pr*PrSH3 plus PrPr*SH3 heterodimer. These are denoted A`, A, B, A2, B2, and AB, respectively. Four equations govern the system (Equations 5–8):
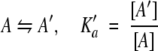 |
(5) |
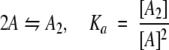 |
(6) |
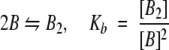 |
(7) |
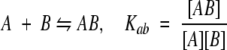 |
(8) |
Secondly, the SH3 occupancy of the dimeric forms of both single mutants is taken to be half that of wild-type PrPrSH3.
The four association constants defined in Equations 5 through 8 are determined through analysis of the distinct association mechanisms of mutant and wild-type fragments. The only association mechanism for Pr*PrSH3 (denoted B) is dimerization without competition from intramolecular self-association, and so the observed association constant for this protein can be used as the value for Kb in Equation 7. The K`a value determined using Equation 4 is used for K`a in Equation 5.
We have already determined KMDa for PrPr*SH3 and PrPrSH3 using a simple monomer-dimer equilibrium model (Equation 2). However, the conformational ensemble of these proteins includes the intramolecular complex in addition to dimer. Therefore, three species are present—intramolecularly bound monomer (A`), free monomer (A), and dimer (A2)—and two equations govern the system (i.e., Equations 5, 6). It can be shown (see Supplementary Material) that the observed association constant (KMDa) obtained when fitting a simple monomer-dimer equilibrium model to a system governed by Equations 5 and 6 is related to K`a and Ka by Equation 9.
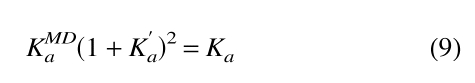 |
(9) |
The observed association constant for PrPr*SH3 may therefore be used to calculate the value of Ka in Equation 6 by use of Equation 9. Similarly, the observed association constant of wild-type PrPrSH3 is used to calculate the value of Kab in Equation 8 using Equation 9 (substituting Kabfor Ka). In so doing, we are assuming that the free energy of asymmetric heterodimer formation (PrPr*SH3 plus Pr*PrSH3) is equal to that of wild-type homodimer.
Including the mass balance for species A and B, we have a set of six equations with six unknowns that can be solved numerically. The equilibrium concentration of all species present can thus be calculated as a function of the total amount of each complimentary mutant in solution. The observed chemical shift for each species can then be computed by using Equation 1, with δPr*Pr*m and δPrPrd substituted for δm and δd, respectively. The fd parameter in Equation 1 thus becomes the mole fraction of occupied to total SH3 pockets giving rise to the NMR signal. For the cross-peak corresponding to Pr*PrSH3, the value of fPr*PrSH3 is calculated using Equation 10:
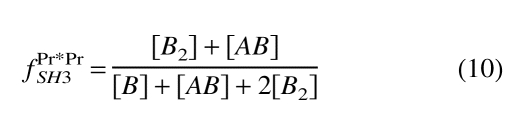 |
(10) |
Similarly, for the cross-peak resulting from the PrPr*SH3 species, fPrPr*SH3 is defined in Equation 11:
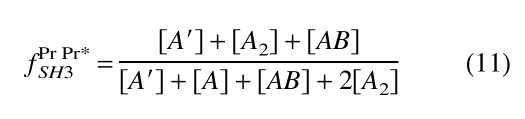 |
(11) |
Each numerator in Equations 10 and 11 is the total molar concentration of occupied SH3 pockets, whereas the denominators are the total molar concentration of SH3 pockets giving rise to the respective signals.
We predicted the concentration-dependent behavior of both cross-peaks in Figure 4C ▶ based only on the results of our initial experiments without adjusting any parameters. The agreement between the predicted and experimental concentration behavior supports the assumptions made in our model. Importantly, the assumption that the free energy of heterodimer formation (Pr*PrSH3 plus PrPr*SH3) is equal to that of wild-type PrPrSH3 homodimer formation is borne out by the accurate prediction of experimental observations.
A priori, it is not evident that the free energy of formation of the heterodimer should equal that of the homodimer. From a statistical mechanical standpoint, we must consider the effect of the distinguishability of the two subunits on the free energy of formation of the heterodimer. Consider a symmetry operation that interconverts the two monomeric subunits of the putative PrPrSH3 dimer as shown in Figure 3E ▶. If we do not distinguish between the two monomeric subunits (i.e., the dimer is a homodimer), then the dimers in Figure 3E ▶ are identical. Therefore, a homodimer has a symmetry number of two (Davidson 1962). If we now consider the monomeric subunit (SH3`) to be distinguishable from the other SH3 (e.g., a residue not involved in mediating dimerization is mutated in SH3`), then the two dimers in Figure 3E ▶ are not identical. The symmetry number of the resulting heterodimer will be one, because no symmetry operation other than identity yields an identical dimer. A consequence of this difference in symmetry numbers is that the association constant of the heterodimer will be twice that of the homodimer (Davidson 1962; Ben-Naim 1992). One might therefore expect the association constant of the heterodimer formed by mixing Pr*PrSH3 and PrPr*SH3 to be twice that of the wild-type PrPrSH3 homodimer.
The heterodimer formed by mixing Pr*PrSH3 and PrPr*SH3 is different from the case described above in that the residues we mutate are involved in mediating dimerization. In the Btk heterodimer (Fig. 3D ▶), two of the four SH3 ligands (proline rich stretches) that mediate dimerization are mutated. As a result, we have eliminated half of the potential stable structures contributing to the association constant of the heterodimer. This cancels out the factor of two in the association constants that would arise because of the different symmetry numbers of the homo- and heterodimers. Thus, the association constants of the Btk hetero- and homodimer will be equal, resulting in equal free energies of formation as assumed in our prediction (Fig. 4C ▶).
The free energies of formation of the homo- and heterodimers may nonetheless differ because of changes in solvation energies or conformational differences that are the result of proline to alanine mutations. However, given the good agreement between predicted and observed experimental values, we believe these differences are negligible. Thus, the data support the wild-type asymmetric self-association model depicted in Figure 3C ▶.
Discussion
Careful investigation of the self-association of the Btk PrPrSH3 fragment has revealed an intricate and novel set of coupled equilibria. The Btk PrPrSH3 polypeptide fragment contains two ligands (proline-rich motifs) and a single ligand-binding surface (SH3 domain). Each of these entities is involved in stabilizing an asymmetric homodimer, in which one of the two SH3 binding pockets in the dimer makes intermolecular contacts to the amino-terminal proline sequence, whereas the other SH3 domain binds to the carboxy-terminal proline motif across the dimer interface. At low concentrations, a monomeric species that involves the amino-terminal proline ligand bound in an intramolecular fashion to the ligand-binding surface of the adjacent SH3 domain predominates. A related protein fragment of the p85 subunit of phosphatidylinositol 3-kinase that contains a single proline motif adjacent to an SH3 domain has also been shown to dimerize in solution but does not show the set of multiple equilibria that we have described here for Btk (Chen and Schreiber 1994). Furthermore, the results presented here for Btk reveal differences among the Tec kinases themselves. The first structural information reported for a Tec kinase involved characterization of the intramolecular complex formed by the proline-rich region of Itk and the neighboring SH3 domain (Andreotti et al. 1997). The most notable difference between Itk and Btk is that Itk self-association is intramolecular, whereas Btk dimerizes and adopts an intramolecular arrangement. Interestingly, the intramolecular complexes formed by Itk and Btk differ. As can be seen in the sequence alignment in Figure 1 ▶, the proline-rich motif of Itk appears to correspond to the carboxy-terminal proline stretch in Btk, yet it is the amino-terminal Btk proline-rich motif that interacts intramolecularly with the Btk SH3 domain. The molecular determinants of the various conformational states of the Tec kinases have yet to be elucidated. Differences in the SH3 domains, the linker regions, and the proline-rich stretches may all contribute to the precise mode of self-association.
The association constants for each of the binding events involving the Btk PrPrSH3 fragment were measured by the combined application of mutagenesis and NMR spectroscopy. The concentration dependence of the mixture of complimentary mutants was accurately predicted by assuming a similar asymmetric association mode to that of wild-type Btk PrPrSH3. It appears that the lower occupancy association modes (Fig. 3B ▶) for the single mutants (PrPr*SH3 and Pr*PrSH3) are not significantly populated in the case of wild-type PrPrSH3. Rather, the Btk PrPrSH3 fragment favors formation of the asymmetric homodimer structure.
The measured association constants indicate that dimerization of Btk PrPrSH3 is a relatively weak interaction. The studies presented here were performed on a small fragment of Btk, and it is conceivable that full-length protein may dimerize via the interactions described here but with greater affinity. In fact, dimerization of full-length Btk has been observed in both gel-filtration assays and mammalian cell systems (X.-Y. Huang, unpubl.). Interestingly, fragments of Itk comprised of the SH3 and SH2 domains also form dimers in solution, albeit via molecular contacts that are distinct from the proline-SH3 interactions described here (Brazin et al. 2000). Together with X-ray crystallographic data that indicate that the PH domain of Btk has a propensity to dimerize (Hyvonen and Saraste 1997), it is becoming apparent that the regulatory domains of the Tec kinases are predisposed under some conditions to form specific dimers.
It is possible that self-association plays a role in regulating Tec family kinase activity. Biochemical investigations into the mechanism of Btk regulation have indicated that PH domain–phosphatidylinositol recognition is responsible for translocating Btk to the plasma membrane where activation proceeds (Kawakami et al. 1994; Li et al. 1997; Bolland et al. 1998; Nisitani et al. 1999). Membrane association is likely accompanied by an increase in the local concentration of Btk that would shift the equilibrium toward dimer. Thus, the intra- and intermolecular species that we have described for the PrPrSH3 fragment of Btk may represent conformational states at distinct points along the activation pathway.
DNA mutations in both proline-rich regions and the SH3 domain of the Btk gene have been detected in XLA patients (Vihinen et al. 1999). Our results show that mutations in either the amino- or carboxy-terminal proline stretches have profoundly different effects on the dominating species in solution. Mutation of the amino-terminal proline sequence (Pr*PrSH3) eliminates the intramolecular interaction. Mutations in the SH3 pocket that reduce the affinity of the domain for proline-rich sequences would be expected to have the same effect as mutation of both proline stretches (Pr*Pr*SH3): elimination of both intra- and intermolecular association. Thus, the competing equilibria (intra- and intermolecular association) involving the PrPrSH3 fragment present a novel system for control of the monomer-dimer equilibrium that could be significantly altered by genetic mutation.
Any exogenous molecule with affinity for either proline sequence or the SH3 domain of Btk (Cheng et al. 1994; Yang et al. 1995) also has the potential to significantly alter the equilibrium species composition of wild-type PrPrSH3. In fact, direct interactions between a heterotrimeric guanine nucleotide-binding protein and the region of Btk that contains the PrPrSH3 sequence studied here result in stimulation of Btk kinase activity (Bence et al. 1997; Ma and Huang 1998). It is also possible that the mechanism of G protein activation is linked to changes in the Btk monomer-dimer equilibrium. As the precise signaling events that control Btk kinase activation are elucidated, it will be important to consider how the intra- and intermolecular modes of self-association presented here contribute to substrate binding and control of catalytic activity.
Materials and methods
Inserts encoding the desired domains of Btk were generated by PCR and were subcloned into the BamHI and EcoRI sites of pGex-2T (Amersham Pharmacia) to generate glutathione-S-transferase (GST) fusion proteins. Domain boundaries of Btk constructs for the Btk SH3 domain were residues 216–275; for the PrPrSH3 domain, residues 175–275. Proline to alanine mutations were introduced using PCR at Pro189 and Pro192 for Pr*PrSH3 and at Pro203 and Pro206 for PrPr*SH3. (All four proline residues are mutated to alanine in the Pr*Pr*SH3 construct.) Protein expression and purification was performed as described previously (Brazin et al. 2000). Buffer conditions for each sample were as follows: 50 mM sodium phosphate (pH 7.4), 150 mM NaCl, 2 mM DTT, 0.02% NaN3 at 25°C. 1H and 15N chemical shifts are referenced to external DSS at 0 ppm (Markley et al. 1998). HSQC spectra were acquired as described (Brazin et al. 2000) on a Bruker DRX 500 spectrometer operating at 499.867 MHz. Fluorescence data were collected using identical buffer conditions as NMR, on an SLM 8100 fluorometer at excitation and emission wavelengths of 295 nm and 430 nm, respectively. Numerical solutions to Equations 5 through 8 and fitting of data to Equation 1 were performed by nonlinear regression using the MATLAB (version 5.3.1, The Mathworks Inc.) suite of programs.
Electronic supplemental material
In the supplemental material, we derive Equation 9 of this manuscript. The file is saved in Adobe PDF format and is called Btksupplemental.pdf.
Acknowledgments
We thank Drs. Jon Applequist, Mark Hargrove, Robert J. Mallis, and Ekaterina Pletneva for fruitful discussions during preparation of this manuscript. In addition, we are grateful to Jya-Wei Cheng (National Tsing Hua University, Hsinchu, Taiwan) for providing coordinates of the SH3-bound peptide. A.L. is supported by a NSF IGERT graduate fellowship award 9972653. This work is supported by a grant from the National Institutes of Health (National Institute of Allergy and Infectious Diseases, AI43957) to A.H.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.26702.
References
- Andreotti, A.H., Bunnell, S.C., Feng, S., Berg, L., and Schreiber, S.L. 1997. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature 385 93–97. [DOI] [PubMed] [Google Scholar]
- Bence, K., Ma, W., Kozasa, T., and Huang, X.Y. 1997. Direct stimulation of Bruton's tyrosine kinase by Gq-protein α-subunit. Nature 389 296–299. [DOI] [PubMed] [Google Scholar]
- Ben-Naim, A. 1992. Statistical thermodynamics for chemists and biochemists. Plenum Press, New York, NY.
- Bolen, J.B. 1995. Protein tyrosine kinases in the initiation of antigen receptor signaling. Curr. Opin. Immunol. 7 306–311. [DOI] [PubMed] [Google Scholar]
- Bolland, S., Pearse, R.N., Kurosaki, T., and Ravetch, J.V. 1998. SHIP modulates immune receptor responses by regulating membrane association of Btk. Immunity 8 509–516. [DOI] [PubMed] [Google Scholar]
- Brazin, K.N., Fulton, D.B., and Andreotti, A.H. 2000. A specific intermolecular association between the regulatory domains of a Tec family kinase. J. Mol. Biol. 302 607–623. [DOI] [PubMed] [Google Scholar]
- Chen, J. and Schreiber, S.L. 1994. SH3 Domain-mediated dimerization of an N-terminal fragment of the phosphatidylinositol 3-kinase p85 subunit Bioorganic and Med. Chem. Lett. 4 1755–1760. [Google Scholar]
- Cheng, G., Ye, Z.-S., and Baltimore, D. 1994. Binding of Bruton's tyrosine kinase to Fyn, Lyn, or Hck through a Src homology 3 domain-mediated interaction. Proc. Natl. Acad. Sci. 91 8152–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J.S. and Shirts, R.B. 1985. Iterative determination of the monomer shift and dimerization constant in a self-associating system J. Phys. Chem. 89 1643–1646. [Google Scholar]
- Davidson, N. 1962. Statistical mechanics. McGraw-Hill, New York, NY.
- Duriez, B. Duquesnoy, P. Dastot, F., Bougneres, P. Amselem, S., and Goossens, M. 1994. An exon-skipping mutation in the btk gene of a patient with X-linked agammaglobulinemia and isolated growth hormone deficiency FEBS Lett. 346 165–170. [DOI] [PubMed] [Google Scholar]
- Feng, S., Chen, J.K., Yu, H., Simon, J.A., and Schreiber, S.L. 1994. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3-ligand interactions. Science 266 1241–1257. [DOI] [PubMed] [Google Scholar]
- Feng, S., Kasahara, C., Rickles, R.J., and Schreiber, S.L. 1995. Specific interactions outside the proline-rich core of two classes of Src homology 3 ligands. Proc. Natl. Acad. Sci. 92 12408–12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson, H., Mattsson, P.T., Allard, P., Haapaniemi, P., Vihinen, M., Smith, C.I.E., and Hard, T. 1998. Solution structure of the SH3 domain from Bruton's tyrosine kinase. Biochemistry 37 2912–2924. [DOI] [PubMed] [Google Scholar]
- Hansson, H., Okoh, M.P., Smith, C.I.E., Vihinen, M., and Hard, T. 2001. Intermolecular interactions between the SH3 domain and the proline-rich TH region of Bruton's tyrosine kinase. FEBS Lett. 489 67–70. [DOI] [PubMed] [Google Scholar]
- Heyeck, S.D. and Berg, L.J. 1993. Developmental regulation of a murine T-cell-specific tyrosine kinase gene, Tsk. Proc. Natl. Acad. Sci. 90 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvonen, M. and Saraste, M. 1997. Structure of the PH domain and Btk motif from Bruton's tyrosine kinase: Molecular explanations for X-linked agammaglobulinaemia. EMBO J. 16 3396–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami, Y., Yao, L., Miura, T., Tsukada, S., Witte, O.N., and Kawakami T. 1994. Tyrosine phosphorylation and activation of Bruton tyrosine kinase upon Fc epsilon RI cross-linking. Mol. Cell. Biol. 14 5108–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. Mol. Graph. 14 29–32. [DOI] [PubMed] [Google Scholar]
- Kuriyan, J. and Cowburn, D. 1997. Modular peptide recognition domains in eukaryotic signaling. Annu. Rev. Biophys. Biomol. Struct. 26 259–288. [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., Ferguson, K.M., and Schlessinger, J. 1996. PH domains: Diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell 85 621–624. [DOI] [PubMed] [Google Scholar]
- Li, Z., Wahl, M.I., Eguinoa, A., Stephens, L.R., Hawkins, P.T., and Witte, O.N. 1997. Phosphatidylinositol 3-kinase-γ activates Bruton's tyrosine kinase in concert with Src family kinases. Proc. Natl. Acad. Sci. 94 13820–13825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Y.C. and Huang, X.Y. 1998. Identification of the binding site for Gqα on its effector Bruton's tyrosine kinase. Proc. Natl. Acad. Sci. 95 12197–12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano, H. 1999. Tec family of protein-tyrosine kinases: An overview of their structure and function.Cytokine Growth Factor Rev. 10 267–280. [DOI] [PubMed] [Google Scholar]
- Markley, J.L., Bax A., Arata, Y., Hilbers, C.W., Kaptein, R., Sykes, B.D., Wright, P.E., and Wütrich, K. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Magn. Reson. 12 1–23. [DOI] [PubMed] [Google Scholar]
- Mori, S., Abeygunawardana, C., O'Neil Johnson, M., and van Zijl, P.C.M. 1995. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J. Magn. Reson. B108 94–98. [DOI] [PubMed] [Google Scholar]
- Nisitani, S., Kato, R.M., Rawlings, D.J., Witte, O.N., and Wahl, M.I. 1999. In situ detection of activated Bruton's tyrosine kinase in the Ig signaling complex by phosphopeptide-specific monoclonal antibodies. Proc. Natl. Acad. Sci. 96 2221–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, Y., Haire, R.N., Litman, R.T., Fu, S.M., Nelson, R.P., Kratz, J., Kornfeld, S.J., Morena, M., Good, R.A., and Litman, G.W. 1994. Genomic organization and structure of Bruton agammaglobulinemia tyrosine kinase: Localization of mutations associated with varied clinical presentations and course in X chromosome–linked agammaglobulinemia. Proc. Natl. Acad. Sci. 91 9062–9066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings, D.J. and Witte, O.N. 1995. The Btk subfamily of cytoplasmic tyrosine kinases: Structure, regulation and function. Semin. Immunol. 7 237–246. [DOI] [PubMed] [Google Scholar]
- Sicheri, F., Moarefi, I., and Kuriyan, J. 1997. Crystal structure of the Src family tyrosine kinase Hck. Nature 385 602–609. [DOI] [PubMed] [Google Scholar]
- Siliciano, J.D., Morrow, T.A., and Desiderio, S.V. 1992. Developmental regulation of a murine T-cell–specific tyrosine kinase gene, Tsk. Proc. Natl. Acad. Sci. 89 11194–11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, S., Saffran, D.C., Rawlings, D.J., Parolini, O., Allen, R.C., Klisak, I., Sparks, R.S., Kubagawa, H., Mohandas, T., Quan, S., Belmont, J.W., Cooper, M.D., Conley, M.E., and Witte, O.N. 1993. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell 72 279–290. [DOI] [PubMed] [Google Scholar]
- Tzeng, S.R., Lou, Y.C., Pai, M.T., Jain, M.L., and Cheng, J.W. 2000. Solution structure of the human BTK SH3 domain complexed with a proline-rich peptide from p120cbl. J. Biomol. NMR 16 303–312. [DOI] [PubMed] [Google Scholar]
- Vetrie, D., Vorechovsky, I., Sideras, P., Holland, J., Davies, A., Flinter, F., Hammarstrom, L., Kinnon, C., Levinsky, R., Bobrow, M., Smith, C.I.E., and Bentley, D.R. 1993. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature 361 226–232. [DOI] [PubMed] [Google Scholar]
- Vihinen, M., Kwan, S.P., Lester, T., Ochs, H.D., Resnick, I., Valiaho, J., Conley, M.E., and Smith, C.I. 1999. Mutations of the human BTK gene coding for Bruton's tyrosine kinase in X-linked agammaglobulinemia. Hum. Mutat. 13 280–285. [DOI] [PubMed] [Google Scholar]
- Vihinen, M., Nilsson, L., and Smith, C.I.E. 1994. Tec homology (TH) adjacent to the PH domain. FEBS Lett. 350 263–265. [DOI] [PubMed] [Google Scholar]
- Xu, W., Harrison, S.C., and Eck, M.J. 1997. Three-dimensional structure of the tyrosine kinase c-Src. Nature 385 595–602. [DOI] [PubMed] [Google Scholar]
- Yang, W. Malek, S.N., and Desiderio, S. 1995. An SH3-binding site conserved in Bruton's tyrosine kinase and related tyrosine kinases mediates specific protein interactions in vitro and in vivo. J. Biol. Chem. 270 20832–20840. [DOI] [PubMed] [Google Scholar]
- Yu, H., Rosen, M.K., Shin, T.B., Seidel-Dugan, C., Brugge, J.S. and Schreiber, S.L. 1992. Solution structure of the SH3 domain of Src and identification of its ligand binding site. Science 258 1665–1668. [DOI] [PubMed] [Google Scholar]