Abstract
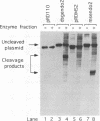
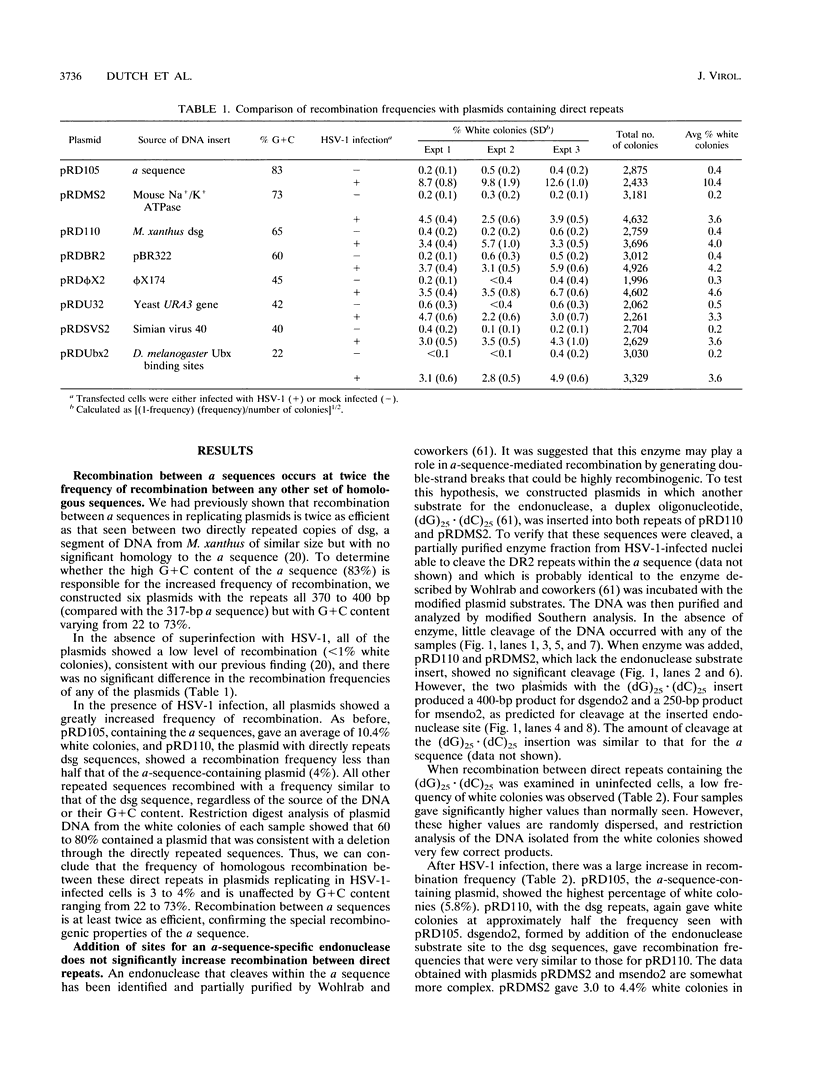
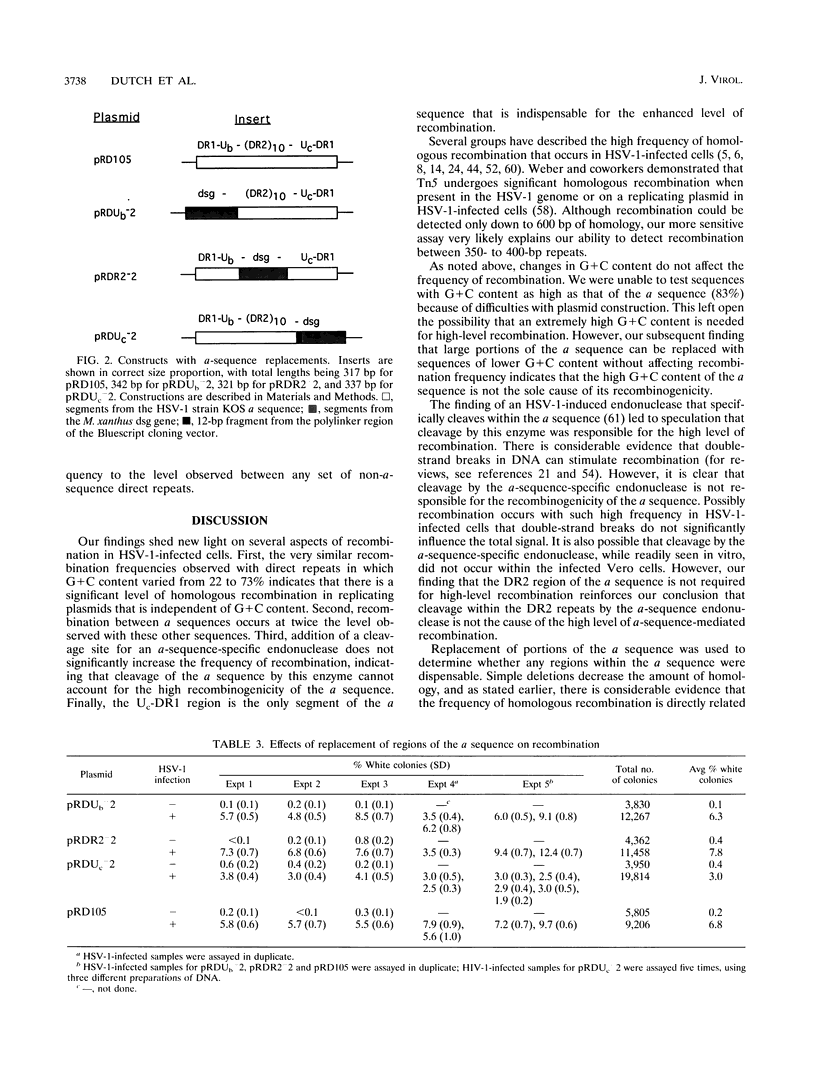
The a sequences of herpes simplex virus type 1 are believed to be the cis sites for inversion events that generate four isomeric forms of the viral genome. Using an assay that measures deletion of a beta-galactosidase gene positioned between two directly repeated sequences in plasmids transiently maintained in Vero cells, we had found that the a sequence is more recombinogenic than another sequence of similar size. To investigate the basis for the enhanced recombination mediated by the a sequence, we examined plasmids containing direct repeats of approximately 350 bp from a variety of sources and with a wide range of G+C content. We observed that all of these plasmids show similar recombination frequencies (3 to 4%) in herpes simplex virus type 1-infected cells. However, recombination between directly repeated a sequences occurs at twice this frequency (6 to 10%). In addition, we find that insertion of a cleavage site for an a-sequence-specific endonuclease into the repeated sequences does not appreciably increase the frequency of recombination, indicating that the presence of endonuclease cleavage sites within the a sequence does not account for its recombinogenicity. Finally, by replacing segments of the a sequence with DNA fragments of similar length, we have determined that only the 95-bp Uc-DR1 segment is indispensable for high-level a-sequence-mediated recombination.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Addison C., Rixon F. J., Palfreyman J. W., O'Hara M., Preston V. G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984 Oct 30;138(2):246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- Addison C., Rixon F. J., Preston V. G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990 Oct;71(Pt 10):2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- Ahn B. Y., Dornfeld K. J., Fagrelius T. J., Livingston D. M. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol Cell Biol. 1988 Jun;8(6):2442–2448. doi: 10.1128/mcb.8.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. M., Ritchie D. A., Subak-Sharpe J. H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973 Mar;18(3):329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- Brown S. M., Subak-Sharpe J. H., Harland J., MacLean A. R. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J Gen Virol. 1992 Feb;73(Pt 2):293–301. doi: 10.1099/0022-1317-73-2-293. [DOI] [PubMed] [Google Scholar]
- Bruckner R. C., Dutch R. E., Zemelman B. V., Mocarski E. S., Lehman I. R. Recombination in vitro between herpes simplex virus type 1 a sequences. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10950–10954. doi: 10.1073/pnas.89.22.10950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubley G. J., Schnipper L. E. Effects of Bloom's syndrome fibroblasts on genetic recombination and mutagenesis of herpes simplex virus type 1. Somat Cell Mol Genet. 1987 Mar;13(2):111–117. doi: 10.1007/BF01534691. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. Characterization of DNA sequence-common and sequence-specific proteins binding to cis-acting sites for cleavage of the terminal a sequence of the herpes simplex virus 1 genome. J Virol. 1989 Mar;63(3):1059–1068. doi: 10.1128/jvi.63.3.1059-1068.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou J., Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985 Jul;41(3):803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- Chou J., Roizman B. The terminal a sequence of the herpes simplex virus genome contains the promoter of a gene located in the repeat sequences of the L component. J Virol. 1986 Feb;57(2):629–637. doi: 10.1128/jvi.57.2.629-637.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu G., Hayakawa H., Berg P. Electroporation for the efficient transfection of mammalian cells with DNA. Nucleic Acids Res. 1987 Feb 11;15(3):1311–1326. doi: 10.1093/nar/15.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalziel R. G., Marsden H. S. Identification of two herpes simplex virus type 1-induced proteins (21K and 22K) which interact specifically with the a sequence of herpes simplex virus DNA. J Gen Virol. 1984 Sep;65(Pt 9):1467–1475. doi: 10.1099/0022-1317-65-9-1467. [DOI] [PubMed] [Google Scholar]
- Dasgupta U. B., Summers W. C. Genetic recombination of herpes simplex virus, the role of the host cell and UV-irradiation of the virus. Mol Gen Genet. 1980;178(3):617–623. doi: 10.1007/BF00337869. [DOI] [PubMed] [Google Scholar]
- Davison A. J., Wilkie N. M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981 Aug;55(Pt 2):315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- Deiss L. P., Chou J., Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986 Sep;59(3):605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss L. P., Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986 Mar;57(3):933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Desai P., DeLuca N. A., Glorioso J. C., Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993 Mar;67(3):1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutch R. E., Bruckner R. C., Mocarski E. S., Lehman I. R. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J Virol. 1992 Jan;66(1):277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E. Exploring the pathways of homologous recombination. Curr Opin Cell Biol. 1992 Jun;4(3):401–412. doi: 10.1016/0955-0674(92)90005-w. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honess R. W., Buchan A., Halliburton I. W., Watson D. H. Recombination and linkage between structural and regulatory genes of herpes simplex virus type 1: study of the functional organization of the genome. J Virol. 1980 Jun;34(3):716–742. doi: 10.1128/jvi.34.3.716-742.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins F. J., Roizman B. Herpes simplex virus 1 recombinants with noninverting genomes frozen in different isomeric arrangements are capable of independent replication. J Virol. 1986 Aug;59(2):494–499. doi: 10.1128/jvi.59.2.494-499.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinks-Robertson S., Michelitch M., Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993 Jul;13(7):3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson F. B., Krasnow M. A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992 Nov;6(11):2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- Kemble G. W., Mocarski E. S. A host cell protein binds to a highly conserved sequence element (pac-2) within the cytomegalovirus a sequence. J Virol. 1989 Nov;63(11):4715–4728. doi: 10.1128/jvi.63.11.4715-4728.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Letsou A., Stachelek J. L. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987 Jan;115(1):161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Deiss L. P., Frenkel N. Nucleotide sequence and structural features of a novel US-a junction present in a defective herpes simplex virus genome. J Virol. 1985 Jul;55(1):140–146. doi: 10.1128/jvi.55.1.140-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Post L. E., Roizman B. Molecular engineering of the herpes simplex virus genome: insertion of a second L-S junction into the genome causes additional genome inversions. Cell. 1980 Nov;22(1 Pt 1):243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Herpesvirus-dependent amplification and inversion of cell-associated viral thymidine kinase gene flanked by viral a sequences and linked to an origin of viral DNA replication. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5626–5630. doi: 10.1073/pnas.79.18.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7047–7051. doi: 10.1073/pnas.78.11.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- Pertuiset B., Boccara M., Cebrian J., Berthelot N., Chousterman S., Puvion-Dutilleul F., Sisman J., Sheldrick P. Physical mapping and nucleotide sequence of a herpes simplex virus type 1 gene required for capsid assembly. J Virol. 1989 May;63(5):2169–2179. doi: 10.1128/jvi.63.5.2169-2179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poffenberger K. L., Tabares E., Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci U S A. 1983 May;80(9):2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue-Geile K. L., Lee G. T., Spear P. G. Novel rearrangements of herpes simplex virus DNA sequences resulting from duplication of a sequence within the unique region of the L component. J Virol. 1985 Feb;53(2):456–461. doi: 10.1128/jvi.53.2.456-461.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Mackem S., Roizman B. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell. 1981 May;24(2):555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- Preston V. G., Coates J. A., Rixon F. J. Identification and characterization of a herpes simplex virus gene product required for encapsidation of virus DNA. J Virol. 1983 Mar;45(3):1056–1064. doi: 10.1128/jvi.45.3.1056-1064.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer P. A., Tevethia M. J., Benyesh-Melnick M. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1974 Mar;58(1):219–228. doi: 10.1016/0042-6822(74)90156-1. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986 Mar;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman G., Bachenheimer S. L. DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology. 1987 Jun;158(2):427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- Singer B. S., Gold L., Gauss P., Doherty D. H. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982 Nov;31(1):25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Duncan J., Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990 Oct;64(10):5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley J. R., Fong B. S., Leung W. C. Construction of a double-jointed herpes simplex viral DNA molecule: inverted repeats are required for segment inversion, and direct repeats promote deletions. Virology. 1981 Aug;113(1):345–362. doi: 10.1016/0042-6822(81)90161-6. [DOI] [PubMed] [Google Scholar]
- Smiley J. R., Lavery C., Howes M. The herpes simplex virus type 1 (HSV-1) a sequence serves as a cleavage/packaging signal but does not drive recombinational genome isomerization when it is inserted into the HSV-2 genome. J Virol. 1992 Dec;66(12):7505–7510. doi: 10.1128/jvi.66.12.7505-7510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stow N. D., Subak-Sharpe J. H., Wilkie N. M. Physical mapping of herpes simplex virus type 1 mutations by marker rescue. J Virol. 1978 Oct;28(1):182–192. doi: 10.1128/jvi.28.1.182-192.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992 Feb;12(2):563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler D. S., Stahl F. W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Umene K. Recombination of the internal direct repeat element DR2 responsible for the fluidity of the a sequence of herpes simplex virus type 1. J Virol. 1991 Oct;65(10):5410–5416. doi: 10.1128/jvi.65.10.5410-5416.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmuza S. L., Smiley J. R. Signals for site-specific cleavage of HSV DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985 Jul;41(3):793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- WILDY P. Recombination with herpes simplex virus. J Gen Microbiol. 1955 Oct;13(2):346–360. doi: 10.1099/00221287-13-2-346. [DOI] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber P. C., Challberg M. D., Nelson N. J., Levine M., Glorioso J. C. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988 Jul 29;54(3):369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- Weber P. C., Levine M., Glorioso J. C. Recombinogenic properties of herpes simplex virus type 1 DNA sequences resident in simian virus 40 minichromosomes. J Virol. 1990 Jan;64(1):300–306. doi: 10.1128/jvi.64.1.300-306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab F., Chatterjee S., Wells R. D. The herpes simplex virus 1 segment inversion site is specifically cleaved by a virus-induced nuclear endonuclease. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6432–6436. doi: 10.1073/pnas.88.15.6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlrab F., McLean M. J., Wells R. D. The segment inversion site of herpes simplex virus type 1 adopts a novel DNA structure. J Biol Chem. 1987 May 5;262(13):6407–6416. [PubMed] [Google Scholar]
- Wohlrab F., Wells R. D. Slight changes in conditions influence the family of non-B-DNA conformations of the herpes simplex virus type 1 DR2 repeats. J Biol Chem. 1989 May 15;264(14):8207–8213. [PubMed] [Google Scholar]
- Yuan L. W., Keil R. L. Distance-independence of mitotic intrachromosomal recombination in Saccharomyces cerevisiae. Genetics. 1990 Feb;124(2):263–273. doi: 10.1093/genetics/124.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al-Kobaisi M. F., Rixon F. J., McDougall I., Preston V. G. The herpes simplex virus UL33 gene product is required for the assembly of full capsids. Virology. 1991 Jan;180(1):380–388. doi: 10.1016/0042-6822(91)90043-b. [DOI] [PubMed] [Google Scholar]