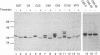
Abstract
Murine polyomavirus contains two related minor coat proteins, VP2 and VP3, in addition to the major coat protein, VP1. The sequence of VP3 is identical to that of the carboxy-terminal two-thirds of VP2. VP2 may serve a role in uncoating of the virus, and both minor coat proteins may be important for viral assembly. In this study, we show that VP3 and a series of deletion mutants of VP3 can be expressed in Escherichia coli as fusion proteins to glutathione S-transferase and partially solubilized with a mild detergent. Using an in vitro binding assay, we demonstrate that a 42-amino-acid fragment near the carboxy terminus of VP3 (residues 140 to 181) is sufficient for binding to purified VP1 pentamers. This binding interaction is rapid, saturable, and specific for the common carboxy terminus of VP2 and VP3. The VP1-VP3 complex can be coimmunoprecipitated with an antibody specific to VP1, and a purified VP3 fragment can selectively extract VP1 from a crude cell lysate. The stoichiometry of the binding reaction suggests that each VP1 pentamer in the virus binds either one VP2 or one VP3, with the VP1-VP2/3 complex stabilized by hydrophobic interactions. These results, taken together with studies from other laboratories on the expression of polyomavirus capsid proteins in mouse and insect cells (S. E. Delos, L. Montross, R. B. Moreland, and R. L. Garcea, Virology, 194:393-398, 1993; J. Forstova, N. Krauzewicz, S. Wallace, A. J. Street, S. M. Dilworth, S. Beard, and B. E. Griffin, J. Virol. 67:1405-1413, 1993), support the idea that a VP1-VP2/3 complex forms in the cytoplasm and, after translocation into the nucleus, acts as the unit for viral assembly.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chou J. Y., Martin R. G. DNA infectivity and the induction of host DNA synthesis with temperature-sensitive mutants of simian virus 40. J Virol. 1975 Jan;15(1):145–150. doi: 10.1128/jvi.15.1.145-150.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow M., Newman J. F., Filman D., Hogle J. M., Rowlands D. J., Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987 Jun 11;327(6122):482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- Clark B., Desselberger U. Myristylation of rotavirus proteins. J Gen Virol. 1988 Oct;69(Pt 10):2681–2686. doi: 10.1099/0022-1317-69-10-2681. [DOI] [PubMed] [Google Scholar]
- Cole C. N., Landers T., Goff S. P., Manteuil-Brutlag S., Berg P. Physical and genetic characterization of deletion mutants of simian virus 40 constructed in vitro. J Virol. 1977 Oct;24(1):277–294. doi: 10.1128/jvi.24.1.277-294.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos S. E., Montross L., Moreland R. B., Garcea R. L. Expression of the polyomavirus VP2 and VP3 proteins in insect cells: coexpression with the major capsid protein VP1 alters VP2/VP3 subcellular localization. Virology. 1993 May;194(1):393–398. doi: 10.1006/viro.1993.1274. [DOI] [PubMed] [Google Scholar]
- Etchison D., Walter G. Subunit interactions in polyoma virus structure. Virology. 1977 Apr;77(2):783–796. doi: 10.1016/0042-6822(77)90499-8. [DOI] [PubMed] [Google Scholar]
- Forstová J., Krauzewicz N., Wallace S., Street A. J., Dilworth S. M., Beard S., Griffin B. E. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J Virol. 1993 Mar;67(3):1405–1413. doi: 10.1128/jvi.67.3.1405-1413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcea R. L., Salunke D. M., Caspar D. L. Site-directed mutation affecting polyomavirus capsid self-assembly in vitro. Nature. 1987 Sep 3;329(6134):86–87. doi: 10.1038/329086a0. [DOI] [PubMed] [Google Scholar]
- Gharakhanian E., Kasamatsu H. Two independent signals, a nuclear localization signal and a Vp1-interactive signal, reside within the carboxy-35 amino acids of SV40 Vp3. Virology. 1990 Sep;178(1):62–71. doi: 10.1016/0042-6822(90)90379-6. [DOI] [PubMed] [Google Scholar]
- Gharakhanian E., Takahashi J., Clever J., Kasamatsu H. In vitro assay for protein-protein interaction: carboxyl-terminal 40 residues of simian virus 40 structural protein VP3 contain a determinant for interaction with VP1. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6607–6611. doi: 10.1073/pnas.85.18.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. P., Griffith D. L., Rayment I., Murakami W. T., Caspar D. L. Inside polyomavirus at 25-A resolution. Nature. 1992 Feb 13;355(6361):652–654. doi: 10.1038/355652a0. [DOI] [PubMed] [Google Scholar]
- Gromeier M., Wetz K. Kinetics of poliovirus uncoating in HeLa cells in a nonacidic environment. J Virol. 1990 Aug;64(8):3590–3597. doi: 10.1128/jvi.64.8.3590-3597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman N., Baltimore D. A plasma membrane component able to bind and alter virions of poliovirus type 1: studies on cell-free alteration using a simplified assay. Virology. 1977 Oct 1;82(1):25–36. doi: 10.1016/0042-6822(77)90029-0. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T., Harrison S. C., Parham P., Brodsky F. M. Location and distribution of the light chains in clathrin trimers. Proc Natl Acad Sci U S A. 1983 May;80(9):2481–2485. doi: 10.1073/pnas.80.9.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauzewicz N., Streuli C. H., Stuart-Smith N., Jones M. D., Wallace S., Griffin B. E. Myristylated polyomavirus VP2: role in the life cycle of the virus. J Virol. 1990 Sep;64(9):4414–4420. doi: 10.1128/jvi.64.9.4414-4420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leavitt A. D., Roberts T. M., Garcea R. L. Polyoma virus major capsid protein, VP1. Purification after high level expression in Escherichia coli. J Biol Chem. 1985 Oct 15;260(23):12803–12809. [PubMed] [Google Scholar]
- Liddington R. C., Yan Y., Moulai J., Sahli R., Benjamin T. L., Harrison S. C. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991 Nov 28;354(6351):278–284. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Lin W., Hata T., Kasamatsu H. Subcellular distribution of viral structural proteins during simian virus 40 infection. J Virol. 1984 May;50(2):363–371. doi: 10.1128/jvi.50.2.363-371.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna R., Xia D., Willingmann P., Ilag L. L., Krishnaswamy S., Rossmann M. G., Olson N. H., Baker T. S., Incardona N. L. Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications. Nature. 1992 Jan 9;355(6356):137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montross L., Watkins S., Moreland R. B., Mamon H., Caspar D. L., Garcea R. L. Nuclear assembly of polyomavirus capsids in insect cells expressing the major capsid protein VP1. J Virol. 1991 Sep;65(9):4991–4998. doi: 10.1128/jvi.65.9.4991-4998.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland R. B., Montross L., Garcea R. L. Characterization of the DNA-binding properties of the polyomavirus capsid protein VP1. J Virol. 1991 Mar;65(3):1168–1176. doi: 10.1128/jvi.65.3.1168-1176.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscufo N., Simons J., Chow M. Myristoylation is important at multiple stages in poliovirus assembly. J Virol. 1991 May;65(5):2372–2380. doi: 10.1128/jvi.65.5.2372-2380.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscufo N., Yafal A. G., Rogove A., Hogle J., Chow M. A mutation in VP4 defines a new step in the late stages of cell entry by poliovirus. J Virol. 1993 Aug;67(8):5075–5078. doi: 10.1128/jvi.67.8.5075-5078.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A. V., Schultz A., Pincus S. E., Oroszlan S., Wimmer E. Capsid protein VP4 of poliovirus is N-myristoylated. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7827–7831. doi: 10.1073/pnas.84.22.7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I., Baker T. S., Caspar D. L., Murakami W. T. Polyoma virus capsid structure at 22.5 A resolution. Nature. 1982 Jan 14;295(5845):110–115. doi: 10.1038/295110a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinemer P., Dirr H. W., Ladenstein R., Schäffer J., Gallay O., Huber R. The three-dimensional structure of class pi glutathione S-transferase in complex with glutathione sulfonate at 2.3 A resolution. EMBO J. 1991 Aug;10(8):1997–2005. doi: 10.1002/j.1460-2075.1991.tb07729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahli R., Freund R., Dubensky T., Garcea R., Bronson R., Benjamin T. Defect in entry and altered pathogenicity of a polyoma virus mutant blocked in VP2 myristylation. Virology. 1993 Jan;192(1):142–153. doi: 10.1006/viro.1993.1016. [DOI] [PubMed] [Google Scholar]
- Salunke D. M., Caspar D. L., Garcea R. L. Polymorphism in the assembly of polyomavirus capsid protein VP1. Biophys J. 1989 Nov;56(5):887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salunke D. M., Caspar D. L., Garcea R. L. Self-assembly of purified polyomavirus capsid protein VP1. Cell. 1986 Sep 12;46(6):895–904. doi: 10.1016/0092-8674(86)90071-1. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Müller H., Schmidt M. F., Rott R. Myristoylation of budgerigar fledgling disease virus capsid protein VP2. J Virol. 1989 Jan;63(1):429–431. doi: 10.1128/jvi.63.1.429-431.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sixma T. K., Pronk S. E., Kalk K. H., Wartna E. S., van Zanten B. A., Witholt B., Hol W. G. Crystal structure of a cholera toxin-related heat-labile enterotoxin from E. coli. Nature. 1991 May 30;351(6325):371–377. doi: 10.1038/351371a0. [DOI] [PubMed] [Google Scholar]
- Sixma T. K., Pronk S. E., Kalk K. H., van Zanten B. A., Berghuis A. M., Hol W. G. Lactose binding to heat-labile enterotoxin revealed by X-ray crystallography. Nature. 1992 Feb 6;355(6360):561–564. doi: 10.1038/355561a0. [DOI] [PubMed] [Google Scholar]
- Smith D. B., Davern K. M., Board P. G., Tiu W. U., Garcia E. G., Mitchell G. F. Mr 26,000 antigen of Schistosoma japonicum recognized by resistant WEHI 129/J mice is a parasite glutathione S-transferase. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8703–8707. doi: 10.1073/pnas.83.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. B., Johnson K. S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988 Jul 15;67(1):31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Streuli C. H., Griffin B. E. Myristic acid is coupled to a structural protein of polyoma virus and SV40. Nature. 1987 Apr 9;326(6113):619–622. doi: 10.1038/326619a0. [DOI] [PubMed] [Google Scholar]