Abstract
E boxes, recognition sequences for basic helix-loop-helix (bHLH) transcription factors, are detected in the enhancer and promoter regions of several murine type C retroviruses. Here we show that ALF1, a member of bHLH protein family of transcription factors, in vitro binds with differing affinities to distinct E-box sequences found in the U3 regulatory regions of Friend, Moloney, SL3-3, and Akv murine leukemia viruses (MLVs) as well as Friend spleen focus-forming virus (SFFV). In NIH 3T3 fibroblasts, ALF1 overexpression elevated transcription from the U3 region of Moloney MLV and the complete long terminal repeat regions of Friend SFFV, Akv MLV, and SL3-3 MLV but neither from the U3 region nor from the complete long terminal repeat of Friend MLV. Introduction of mutations in the Akv MLV E boxes showed the E-box cis elements to be required for the function of ALF1 as a transcription factor. ALF1 and the glucocorticoid receptor, with overlapping DNA binding sequences, did not act synergistically with respect to transcriptional trans activation of expression from the Akv MLV promoter-enhancer region. We conclude that ALF1 in vivo may be an important transcription regulator for Akv, SL3-3, and Moloney MLVs as well as for Friend SFFV.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews N. C., Faller D. V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991 May 11;19(9):2499–2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronheim A., Ohlsson H., Park C. W., Edlund T., Walker M. D. Distribution and characterization of helix-loop-helix enhancer-binding proteins from pancreatic beta cells and lymphocytes. Nucleic Acids Res. 1991 Jul 25;19(14):3893–3899. doi: 10.1093/nar/19.14.3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990 Apr 6;61(1):49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Kretzner L., Blackwood E. M., Eisenman R. N., Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990 Nov 23;250(4984):1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Celander D., Haseltine W. A. Tissue-specific transcription preference as a determinant of cell tropism and leukaemogenic potential of murine retroviruses. Nature. 1984 Nov 8;312(5990):159–162. doi: 10.1038/312159a0. [DOI] [PubMed] [Google Scholar]
- Celander D., Hsu B. L., Haseltine W. A. Regulatory elements within the murine leukemia virus enhancer regions mediate glucocorticoid responsiveness. J Virol. 1988 Apr;62(4):1314–1322. doi: 10.1128/jvi.62.4.1314-1322.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Hartley J. W., Rowe W. P., Hopkins N. Role for the 3' end of the genome in determining disease specificity of Friend and Moloney murine leukemia viruses. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4408–4411. doi: 10.1073/pnas.80.14.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatis P. A., Holland C. A., Silver J. E., Frederickson T. N., Hopkins N., Hartley J. W. A 3' end fragment encompassing the transcriptional enhancers of nondefective Friend virus confers erythroleukemogenicity on Moloney leukemia virus. J Virol. 1984 Oct;52(1):248–254. doi: 10.1128/jvi.52.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy B. A., Sanders L. K., Lau L. F., Copeland N. G., Jenkins N. A., Nathans D. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1815–1819. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. P., Mak T. W. Nucleotide sequences of the murine retrovirus Friend SFFVp long terminal repeats: identification of a structure with extensive dyad symmetry 5' to the TATA box. Nucleic Acids Res. 1982 May 25;10(10):3315–3330. doi: 10.1093/nar/10.10.3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneliussen B., Thornell A., Hallberg B., Grundström T. Helix-loop-helix transcriptional activators bind to a sequence in glucocorticoid response elements of retrovirus enhancers. J Virol. 1991 Nov;65(11):6084–6093. doi: 10.1128/jvi.65.11.6084-6093.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Jolicoeur P. The tandem direct repeats within the long terminal repeat of murine leukemia viruses are the primary determinant of their leukemogenic potential. J Virol. 1984 Dec;52(3):945–952. doi: 10.1128/jvi.52.3.945-952.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesGroseillers L., Villemur R., Jolicoeur P. The high leukemogenic potential of Gross passage A murine leukemia virus maps in the region of the genome corresponding to the long terminal repeat and to the 3' end of env. J Virol. 1983 Jul;47(1):24–32. doi: 10.1128/jvi.47.1.24-32.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duch M., Paludan K., Lovmand J., Pedersen L., Jørgensen P., Pedersen F. S. A correlation between dexamethasone inducibility and basal expression levels of retroviral vector proviruses. Nucleic Acids Res. 1993 Oct 11;21(20):4777–4782. doi: 10.1093/nar/21.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis E. A., Speck N. A., Hopkins N. Alignment of U3 region sequences of mammalian type C viruses: identification of highly conserved motifs and implications for enhancer design. J Virol. 1990 Feb;64(2):534–542. doi: 10.1128/jvi.64.2.534-542.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C. V., Jacob P. E., Ringold G. M., Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2(1):101–109. [PubMed] [Google Scholar]
- Hallberg B., Schmidt J., Luz A., Pedersen F. S., Grundström T. SL3-3 enhancer factor 1 transcriptional activators are required for tumor formation by SL3-3 murine leukemia virus. J Virol. 1991 Aug;65(8):4177–4181. doi: 10.1128/jvi.65.8.4177-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Henthorn P., Kiledjian M., Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990 Jan 26;247(4941):467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- Hsu H. L., Huang L., Tsan J. T., Funk W., Wright W. E., Hu J. S., Kingston R. E., Baer R. Preferred sequences for DNA recognition by the TAL1 helix-loop-helix proteins. Mol Cell Biol. 1994 Feb;14(2):1256–1265. doi: 10.1128/mcb.14.2.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. S., Olson E. N., Kingston R. E. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA-binding ability of myogenic regulatory factors. Mol Cell Biol. 1992 Mar;12(3):1031–1042. doi: 10.1128/mcb.12.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto A., Takimoto M., Adachi A., Kakuyama M., Kato S., Kakimi K., Fukuoka K., Ogiu T., Matsuyama M. Sequences responsible for erythroid and lymphoid leukemia in the long terminal repeats of Friend-mink cell focus-forming and Moloney murine leukemia viruses. J Virol. 1987 Jun;61(6):1861–1866. doi: 10.1128/jvi.61.6.1861-1866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Y., Vierra C., Nelson C. E2A expression, nuclear localization, and in vivo formation of DNA- and non-DNA-binding species during B-cell development. Mol Cell Biol. 1993 Dec;13(12):7321–7333. doi: 10.1128/mcb.13.12.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski M., Merregaert J., Boniver J., Maisin J. R. Proviral genome of radiation leukemia virus: molecular cloning of biologically active proviral DNA and nucleotide sequence of its long terminal repeat. J Virol. 1985 Jul;55(1):251–255. doi: 10.1128/jvi.55.1.251-255.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jen Y., Weintraub H., Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: in vivo association of Id with E2A proteins. Genes Dev. 1992 Aug;6(8):1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- Klein E. S., Simmons D. M., Swanson L. W., Rosenfeld M. G. Tissue-specific RNA splicing generates an ankyrin-like domain that affects the dimerization and DNA-binding properties of a bHLH protein. Genes Dev. 1993 Jan;7(1):55–71. doi: 10.1101/gad.7.1.55. [DOI] [PubMed] [Google Scholar]
- Koch W., Zimmermann W., Oliff A., Friedrich R. Molecular analysis of the envelope gene and long terminal repeat of Friend mink cell focus-inducing virus: implications for the functions of these sequences. J Virol. 1984 Mar;49(3):828–840. doi: 10.1128/jvi.49.3.828-840.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassar A. B., Davis R. L., Wright W. E., Kadesch T., Murre C., Voronova A., Baltimore D., Weintraub H. Functional activity of myogenic HLH proteins requires hetero-oligomerization with E12/E47-like proteins in vivo. Cell. 1991 Jul 26;66(2):305–315. doi: 10.1016/0092-8674(91)90620-e. [DOI] [PubMed] [Google Scholar]
- Lenardo M., Pierce J. W., Baltimore D. Protein-binding sites in Ig gene enhancers determine transcriptional activity and inducibility. Science. 1987 Jun 19;236(4808):1573–1577. doi: 10.1126/science.3109035. [DOI] [PubMed] [Google Scholar]
- Lenz J., Celander D., Crowther R. L., Patarca R., Perkins D. W., Haseltine W. A. Determination of the leukaemogenicity of a murine retrovirus by sequences within the long terminal repeat. 1984 Mar 29-Apr 4Nature. 308(5958):467–470. doi: 10.1038/308467a0. [DOI] [PubMed] [Google Scholar]
- Lenz J., Crowther R., Klimenko S., Haseltine W. Molecular cloning of a highly leukemogenic, ecotropic retrovirus from an AKR mouse. J Virol. 1982 Sep;43(3):943–951. doi: 10.1128/jvi.43.3.943-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Golemis E., Hartley J. W., Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987 Mar;61(3):693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linemeyer D. L., Menke J. G., Ruscetti S. K., Evans L. H., Scolnick E. M. Envelope gene sequences which encode the gp52 protein of spleen focus-forming virus are required for the induction of erythroid cell proliferation. J Virol. 1982 Jul;43(1):223–233. doi: 10.1128/jvi.43.1.223-233.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovmand S., Kjeldgaard N. O., Jørgensen P., Pedersen F. S. Enhancer functions in U3 of Akv virus: a role for cooperativity of a tandem repeat unit and its flanking DNA sequences. J Virol. 1990 Jul;64(7):3185–3191. doi: 10.1128/jvi.64.7.3185-3191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N. R., O'Connell M. A., Sharp P. A., Hopkins N. Nuclear factors that bind to the enhancer region of nondefective Friend murine leukemia virus. J Virol. 1989 Oct;63(10):4210–4223. doi: 10.1128/jvi.63.10.4210-4223.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley N. R., O'Connell M., Sun W., Speck N. A., Hopkins N. Two factors that bind to highly conserved sequences in mammalian type C retroviral enhancers. J Virol. 1993 Apr;67(4):1967–1975. doi: 10.1128/jvi.67.4.1967-1975.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miksicek R., Heber A., Schmid W., Danesch U., Posseckert G., Beato M., Schütz G. Glucocorticoid responsiveness of the transcriptional enhancer of Moloney murine sarcoma virus. Cell. 1986 Jul 18;46(2):283–290. doi: 10.1016/0092-8674(86)90745-2. [DOI] [PubMed] [Google Scholar]
- Mizushima S., Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990 Sep 11;18(17):5322–5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989 Mar 10;56(5):777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989 Aug 11;58(3):537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Murre C., Voronova A., Baltimore D. B-cell- and myocyte-specific E2-box-binding factors contain E12/E47-like subunits. Mol Cell Biol. 1991 Feb;11(2):1156–1160. doi: 10.1128/mcb.11.2.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelsen B., Tian G., Erman B., Gregoire J., Maki R., Graves B., Sen R. Regulation of lymphoid-specific immunoglobulin mu heavy chain gene enhancer by ETS-domain proteins. Science. 1993 Jul 2;261(5117):82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- Nelson C., Shen L. P., Meister A., Fodor E., Rutter W. J. Pan: a transcriptional regulator that binds chymotrypsin, insulin, and AP-4 enhancer motifs. Genes Dev. 1990 Jun;4(6):1035–1043. doi: 10.1101/gad.4.6.1035. [DOI] [PubMed] [Google Scholar]
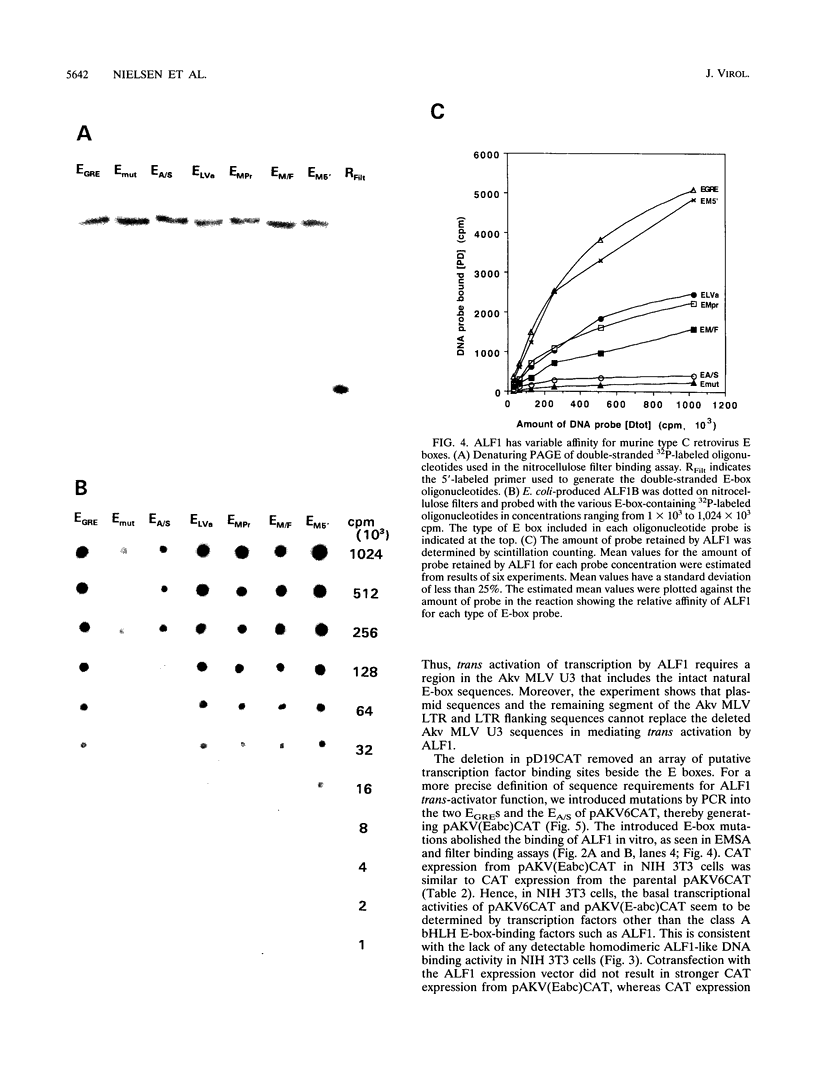
- Nielsen A. L., Pallisgaard N., Pedersen F. S., Jørgensen P. Murine helix-loop-helix transcriptional activator proteins binding to the E-box motif of the Akv murine leukemia virus enhancer identified by cDNA cloning. Mol Cell Biol. 1992 Aug;12(8):3449–3459. doi: 10.1128/mcb.12.8.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nørby P. L., Pallisgaard N., Pedersen F. S., Jørgensen P. Determination of recognition-sequences for DNA-binding proteins by a polymerase chain reaction assisted binding site selection method (BSS) using nitrocellulose immobilized DNA binding protein. Nucleic Acids Res. 1992 Dec 11;20(23):6317–6321. doi: 10.1093/nar/20.23.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen H. S., Lovmand S., Lovmand J., Jørgensen P., Kjeldgaard N. O., Pedersen F. S. Involvement of nuclear factor I-binding sites in control of Akv virus gene expression. J Virol. 1990 Sep;64(9):4152–4161. doi: 10.1128/jvi.64.9.4152-4161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallisgaard N., Pedersen F. S., Birkelund S., Jørgensen P. A common multiple cloning site in a set of vectors for expression of eukaryotic genes in mammalian, insect and bacterial cells. Gene. 1994 Jan 28;138(1-2):115–118. doi: 10.1016/0378-1119(94)90791-9. [DOI] [PubMed] [Google Scholar]
- Paludan K., Dai H. Y., Duch M., Jørgensen P., Kjeldgaard N. O., Pedersen F. S. Different relative expression from two murine leukemia virus long terminal repeats in unintegrated transfected DNA and in integrated retroviral vector proviruses. J Virol. 1989 Dec;63(12):5201–5207. doi: 10.1128/jvi.63.12.5201-5207.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongubala J. M., Atchison M. L. Functional characterization of the developmentally controlled immunoglobulin kappa 3' enhancer: regulation by Id, a repressor of helix-loop-helix transcription factors. Mol Cell Biol. 1991 Feb;11(2):1040–1047. doi: 10.1128/mcb.11.2.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera R. R., Stuiver M. H., Steenbergen R., Murre C. Ets proteins: new factors that regulate immunoglobulin heavy-chain gene expression. Mol Cell Biol. 1993 Nov;13(11):7163–7169. doi: 10.1128/mcb.13.11.7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen C. A., Haseltine W. A., Lenz J., Ruprecht R., Cloyd M. W. Tissue selectivity of murine leukemia virus infection is determined by long terminal repeat sequences. J Virol. 1985 Sep;55(3):862–866. doi: 10.1128/jvi.55.3.862-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruta M., Bestwick R., Machida C., Kabat D. Loss of leukemogenicity caused by mutations in the membrane glycoprotein structural gene of Friend spleen focus-forming virus. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4704–4708. doi: 10.1073/pnas.80.15.4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S., Littman D. R. A heterodimer of HEB and an E12-related protein interacts with the CD4 enhancer and regulates its activity in T-cell lines. Mol Cell Biol. 1993 Sep;13(9):5620–5628. doi: 10.1128/mcb.13.9.5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Short M. K., Okenquist S. A., Lenz J. Correlation of leukemogenic potential of murine retroviruses with transcriptional tissue preference of the viral long terminal repeats. J Virol. 1987 Apr;61(4):1067–1072. doi: 10.1128/jvi.61.4.1067-1072.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Baltimore D. Six distinct nuclear factors interact with the 75-base-pair repeat of the Moloney murine leukemia virus enhancer. Mol Cell Biol. 1987 Mar;7(3):1101–1110. doi: 10.1128/mcb.7.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Golemis E., Fredrickson T. N., Hartley J. W., Hopkins N. Mutation of the core or adjacent LVb elements of the Moloney murine leukemia virus enhancer alters disease specificity. Genes Dev. 1990 Feb;4(2):233–242. doi: 10.1101/gad.4.2.233. [DOI] [PubMed] [Google Scholar]
- Speck N. A., Renjifo B., Hopkins N. Point mutations in the Moloney murine leukemia virus enhancer identify a lymphoid-specific viral core motif and 1,3-phorbol myristate acetate-inducible element. J Virol. 1990 Feb;64(2):543–550. doi: 10.1128/jvi.64.2.543-550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro C., Li J. P., Bestwick R. K., Kabat D. An enhancer sequence instability that diversifies the cell repertoire for expression of a murine leukemia virus. Virology. 1988 Jun;164(2):350–361. doi: 10.1016/0042-6822(88)90548-x. [DOI] [PubMed] [Google Scholar]
- Sun X. H., Baltimore D. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell. 1991 Jan 25;64(2):459–470. doi: 10.1016/0092-8674(91)90653-g. [DOI] [PubMed] [Google Scholar]
- Sun X. H., Copeland N. G., Jenkins N. A., Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol Cell Biol. 1991 Nov;11(11):5603–5611. doi: 10.1128/mcb.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. D., Park C. W., Rosen A., Aronheim A. A cDNA from a mouse pancreatic beta cell encoding a putative transcription factor of the insulin gene. Nucleic Acids Res. 1990 Mar 11;18(5):1159–1166. doi: 10.1093/nar/18.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Dwarki V. J., Verma I., Davis R., Hollenberg S., Snider L., Lassar A., Tapscott S. J. Muscle-specific transcriptional activation by MyoD. Genes Dev. 1991 Aug;5(8):1377–1386. doi: 10.1101/gad.5.8.1377. [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Kiledjian M., Shen C. P., Benezra R., Zwollo P., Dymecki S. M., Desiderio S. V., Kadesch T. Repression of immunoglobulin enhancers by the helix-loop-helix protein Id: implications for B-lymphoid-cell development. Mol Cell Biol. 1991 Dec;11(12):6185–6191. doi: 10.1128/mcb.11.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff L., Kaminchik J., Hankins W. D., Ruscetti S. K. Sequence comparisons of the anemia- and polycythemia-inducing strains of Friend spleen focus-forming virus. J Virol. 1985 Feb;53(2):570–578. doi: 10.1128/jvi.53.2.570-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol. 1966 Mar;16(1):118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- Wotton D., Ghysdael J., Wang S., Speck N. A., Owen M. J. Cooperative binding of Ets-1 and core binding factor to DNA. Mol Cell Biol. 1994 Jan;14(1):840–850. doi: 10.1128/mcb.14.1.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Babin J., Feldhaus A. L., Singh H., Sharp P. A., Bina M. HTF4: a new human helix-loop-helix protein. Nucleic Acids Res. 1991 Aug 25;19(16):4555–4555. doi: 10.1093/nar/19.16.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Doyle K., Bina M. Interactions of HTF4 with E-box motifs in the long terminal repeat of human immunodeficiency virus type 1. J Virol. 1992 Sep;66(9):5631–5634. doi: 10.1128/jvi.66.9.5631-5634.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]