Abstract
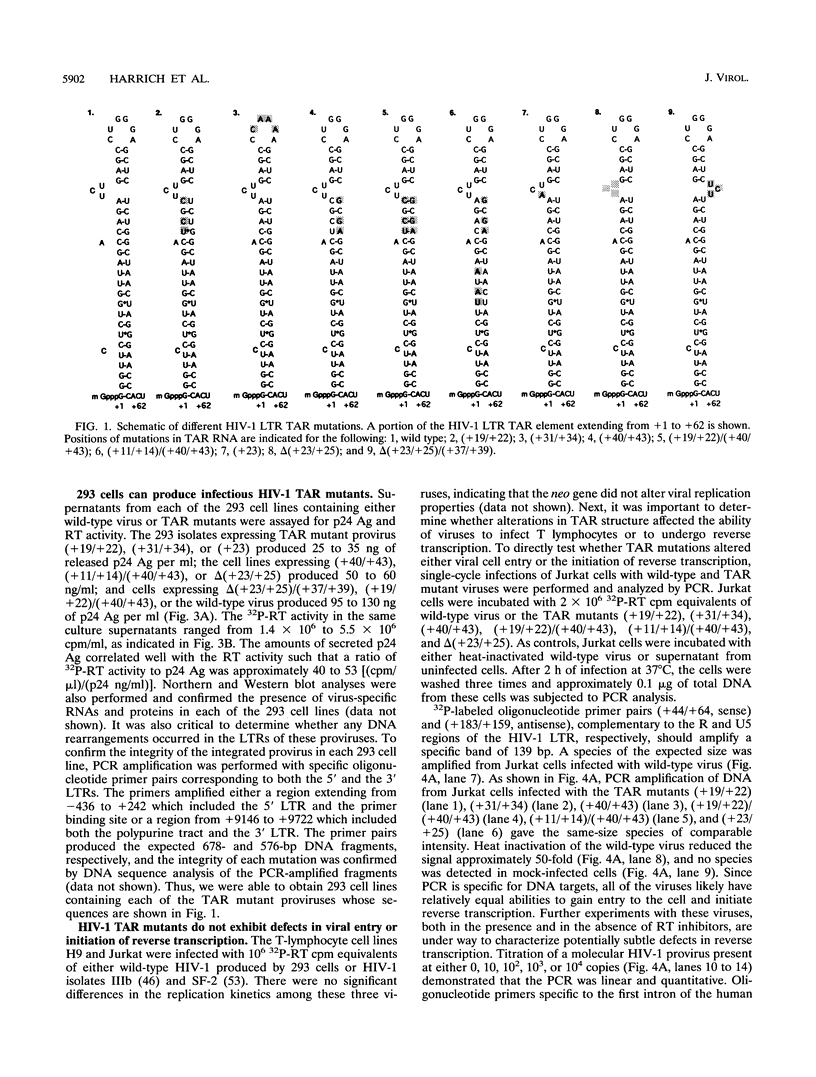
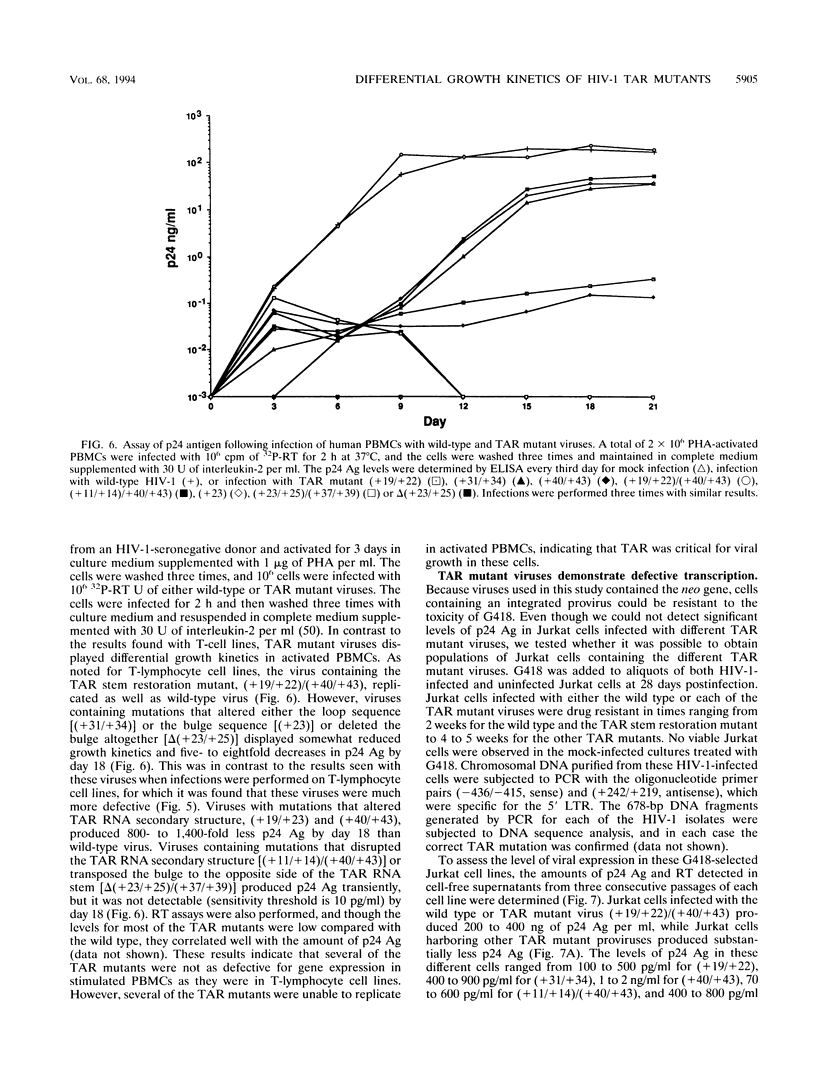
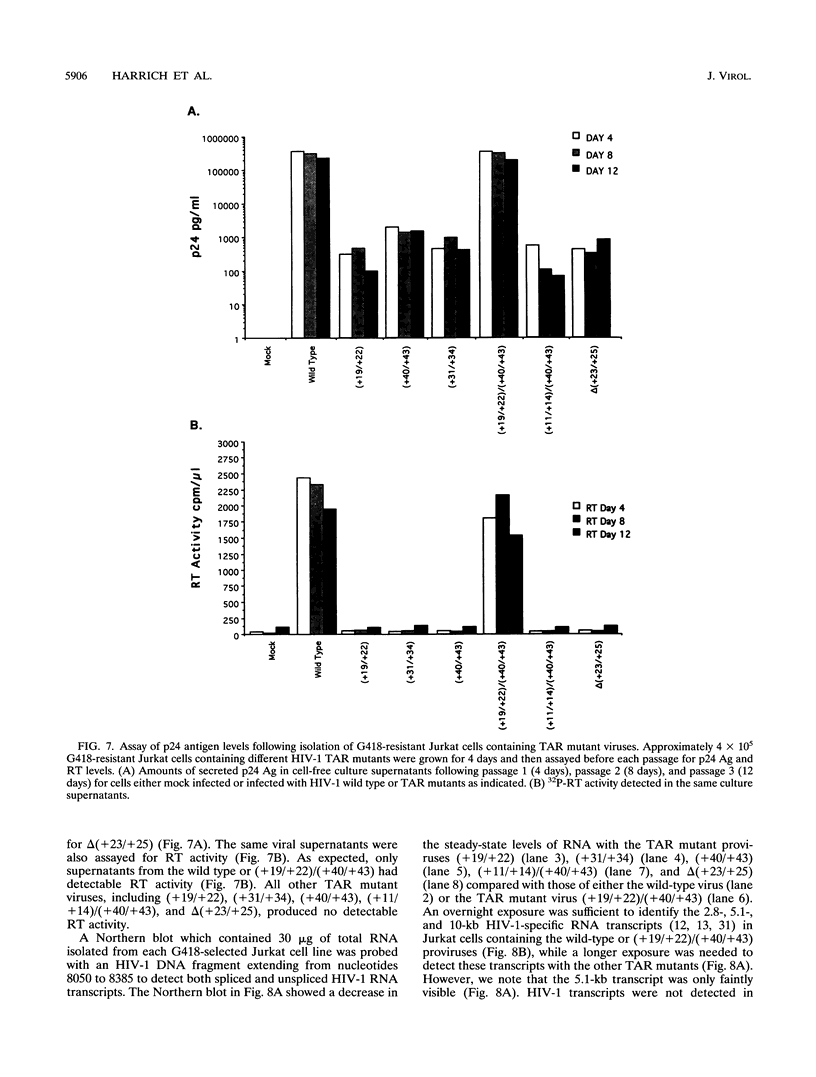
The human immunodeficiency virus type 1 (HIV-1) TAR element is critical for the activation of gene expression by the transactivator protein, Tat. Mutagenesis has demonstrated that a stable stem-loop RNA structure containing both loop and bulge structures transcribed from TAR is the major target for tat activation. Though transient assays have defined elements critical for TAR function, no studies have yet determined the role of TAR in viral replication because of the inability to generate viral stocks containing mutations in TAR. In the current study, we developed a strategy which enabled us to generate stable 293 cell lines which were capable of producing high titers of different viruses containing TAR mutations. Viruses generated from these cell lines were used to infect both T-lymphocyte cell lines and peripheral blood mononuclear cells. Viruses containing TAR mutations in either the upper stem, the bulge, or the loop exhibited dramatically decreased HIV-1 gene expression and replication in all cell lines tested. However, we were able to isolate lymphoid cell lines which stably expressed gene products from each of these TAR mutant viruses. Though the amounts of virus in these cell lines were roughly equivalent, cells containing TAR mutant viruses were extremely defective for gene expression compared with cell lines containing wild-type virus. The magnitude of this decrease in viral gene expression was much greater than previously seen in transient expression assays using HIV-1 long terminal repeat chloramphenicol acetyltransferase gene constructs. In contrast to the defects in viral growth found in T-lymphocyte cell lines, several of the viruses containing TAR mutations were much less defective for gene expression and replication in activated peripheral blood mononuclear cells. These results indicate that maintenance of the TAR element is critical for viral gene expression and replication in all cell lines tested, though the cell type which is infected is also a major determinant of the replication properties of TAR mutant viruses.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barry P. A., Pratt-Lowe E., Unger R. E., Luciw P. A. Cellular factors regulate transactivation of human immunodeficiency virus type 1. J Virol. 1991 Mar;65(3):1392–1399. doi: 10.1128/jvi.65.3.1392-1399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Gatignol A., Rabson A. B., Jeang K. T. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990 Aug 24;62(4):757–767. doi: 10.1016/0092-8674(90)90120-4. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1992 Jan;66(1):139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. trans activation of human immunodeficiency virus type 1 is sequence specific for both the single-stranded bulge and loop of the trans-acting-responsive hairpin: a quantitative analysis. J Virol. 1989 Dec;63(12):5501–5504. doi: 10.1128/jvi.63.12.5501-5504.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Caputo A., Sodroski J. G., Haseltine W. A. Constitutive expression of HIV-1 tat protein in human Jurkat T cells using a BK virus vector. J Acquir Immune Defic Syndr. 1990;3(4):372–379. [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayton A. I., Sodroski J. G., Rosen C. A., Goh W. C., Haseltine W. A. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986 Mar 28;44(6):941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990 Dec;9(12):4145–4153. doi: 10.1002/j.1460-2075.1990.tb07637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M. B., Baltimore D., Frankel A. D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci U S A. 1991 May 1;88(9):4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M. B., Jarrett R. F., Aldovini A., Gallo R. C., Wong-Staal F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell. 1986 Sep 12;46(6):807–817. doi: 10.1016/0092-8674(86)90062-0. [DOI] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Fisher A. G., Feinberg M. B., Josephs S. F., Harper M. E., Marselle L. M., Reyes G., Gonda M. A., Aldovini A., Debouk C., Gallo R. C. The trans-activator gene of HTLV-III is essential for virus replication. 1986 Mar 27-Apr 2Nature. 320(6060):367–371. doi: 10.1038/320367a0. [DOI] [PubMed] [Google Scholar]
- Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., Gaynor R. B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989 Mar;8(3):765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J. A., Wu F. K., Mitsuyasu R., Gaynor R. B. Interactions of cellular proteins involved in the transcriptional regulation of the human immunodeficiency virus. EMBO J. 1987 Dec 1;6(12):3761–3770. doi: 10.1002/j.1460-2075.1987.tb02711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991 Mar 29;251(5001):1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Gaynor R. Cellular transcription factors involved in the regulation of HIV-1 gene expression. AIDS. 1992 Apr;6(4):347–363. doi: 10.1097/00002030-199204000-00001. [DOI] [PubMed] [Google Scholar]
- Graeble M. A., Churcher M. J., Lowe A. D., Gait M. J., Karn J. Human immunodeficiency virus type 1 transactivator protein, tat, stimulates transcriptional read-through of distal terminator sequences in vitro. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6184–6188. doi: 10.1073/pnas.90.13.6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., Smiley J., Russell W. C., Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977 Jul;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Harrich D., Garcia J., Mitsuyasu R., Gaynor R. TAR independent activation of the human immunodeficiency virus in phorbol ester stimulated T lymphocytes. EMBO J. 1990 Dec;9(13):4417–4423. doi: 10.1002/j.1460-2075.1990.tb07892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrich D., Garcia J., Wu F., Mitsuyasu R., Gonazalez J., Gaynor R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1989 Jun;63(6):2585–2591. doi: 10.1128/jvi.63.6.2585-2591.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauber J., Cullen B. R. Mutational analysis of the trans-activation-responsive region of the human immunodeficiency virus type I long terminal repeat. J Virol. 1988 Mar;62(3):673–679. doi: 10.1128/jvi.62.3.673-679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K. A., Kadonaga J. T., Luciw P. A., Tjian R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science. 1986 May 9;232(4751):755–759. doi: 10.1126/science.3008338. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Luciw P. A., Duchange N. Structural arrangements of transcription control domains within the 5'-untranslated leader regions of the HIV-1 and HIV-2 promoters. Genes Dev. 1988 Sep;2(9):1101–1114. doi: 10.1101/gad.2.9.1101. [DOI] [PubMed] [Google Scholar]
- Kao S. Y., Calman A. F., Luciw P. A., Peterlin B. M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987 Dec 3;330(6147):489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- Kato H., Horikoshi M., Roeder R. G. Repression of HIV-1 transcription by a cellular protein. Science. 1991 Mar 22;251(5000):1476–1479. doi: 10.1126/science.2006421. [DOI] [PubMed] [Google Scholar]
- Kato H., Sumimoto H., Pognonec P., Chen C. H., Rosen C. A., Roeder R. G. HIV-1 Tat acts as a processivity factor in vitro in conjunction with cellular elongation factors. Genes Dev. 1992 Apr;6(4):655–666. doi: 10.1101/gad.6.4.655. [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Byrn R., Groopman J., Baltimore D. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J Virol. 1989 Sep;63(9):3708–3713. doi: 10.1128/jvi.63.9.3708-3713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S., Garcia J., Pearson L., Soultanakis E., Dasgupta A., Gaynor R. Multiple transcriptional regulatory domains in the human immunodeficiency virus type 1 long terminal repeat are involved in basal and E1A/E1B-induced promoter activity. J Virol. 1989 Nov;63(11):4616–4625. doi: 10.1128/jvi.63.11.4616-4625.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laspia M. F., Rice A. P., Mathews M. B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989 Oct 20;59(2):283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- Laspia M. F., Rice A. P., Mathews M. B. Synergy between HIV-1 Tat and adenovirus E1A is principally due to stabilization of transcriptional elongation. Genes Dev. 1990 Dec;4(12B):2397–2408. doi: 10.1101/gad.4.12b.2397. [DOI] [PubMed] [Google Scholar]
- Leonard J., Parrott C., Buckler-White A. J., Turner W., Ross E. K., Martin M. A., Rabson A. B. The NF-kappa B binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989 Nov;63(11):4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Welsh T. M., Peterlin B. M. The human immunodeficiency virus type 1 long terminal repeat specifies two different transcription complexes, only one of which is regulated by Tat. J Virol. 1993 Apr;67(4):1752–1760. doi: 10.1128/jvi.67.4.1752-1760.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Stenzel M., Sodroski J. G., Haseltine W. A. Effects of long terminal repeat mutations on human immunodeficiency virus type 1 replication. J Virol. 1989 Sep;63(9):4115–4119. doi: 10.1128/jvi.63.9.4115-4119.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R. A., Sharp P. A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991 Dec;10(13):4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Nabel G. J., Rice S. A., Knipe D. M., Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988 Mar 11;239(4845):1299–1302. doi: 10.1126/science.2830675. [DOI] [PubMed] [Google Scholar]
- Nabel G., Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987 Apr 16;326(6114):711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Okamoto T., Wong-Staal F. Demonstration of virus-specific transcriptional activator(s) in cells infected with HTLV-III by an in vitro cell-free system. Cell. 1986 Oct 10;47(1):29–35. doi: 10.1016/0092-8674(86)90363-6. [DOI] [PubMed] [Google Scholar]
- Olsen H. S., Rosen C. A. Contribution of the TATA motif to Tat-mediated transcriptional activation of human immunodeficiency virus gene expression. J Virol. 1992 Sep;66(9):5594–5597. doi: 10.1128/jvi.66.9.5594-5597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott C., Seidner T., Duh E., Leonard J., Theodore T. S., Buckler-White A., Martin M. A., Rabson A. B. Variable role of the long terminal repeat Sp1-binding sites in human immunodeficiency virus replication in T lymphocytes. J Virol. 1991 Mar;65(3):1414–1419. doi: 10.1128/jvi.65.3.1414-1419.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin B. M., Luciw P. A., Barr P. J., Walker M. D. Elevated levels of mRNA can account for the trans-activation of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9734–9738. doi: 10.1073/pnas.83.24.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Sarngadharan M. G., Read E., Gallo R. C. Detection, isolation, and continuous production of cytopathic retroviruses (HTLV-III) from patients with AIDS and pre-AIDS. Science. 1984 May 4;224(4648):497–500. doi: 10.1126/science.6200935. [DOI] [PubMed] [Google Scholar]
- Ratnasabapathy R., Sheldon M., Johal L., Hernandez N. The HIV-1 long terminal repeat contains an unusual element that induces the synthesis of short RNAs from various mRNA and snRNA promoters. Genes Dev. 1990 Dec;4(12A):2061–2074. doi: 10.1101/gad.4.12a.2061. [DOI] [PubMed] [Google Scholar]
- Rosen C. A., Sodroski J. G., Haseltine W. A. Location of cis-acting regulatory sequences in the human T-cell leukemia virus type I long terminal repeat. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6502–6506. doi: 10.1073/pnas.82.19.6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. K., Buckler-White A. J., Rabson A. B., Englund G., Martin M. A. Contribution of NF-kappa B and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J Virol. 1991 Aug;65(8):4350–4358. doi: 10.1128/jvi.65.8.4350-4358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Roy S., Parkin N. T., Rosen C., Itovitch J., Sonenberg N. Structural requirements for trans activation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by tat: importance of base pairing, loop sequence, and bulges in the tat-responsive sequence. J Virol. 1990 Mar;64(3):1402–1406. doi: 10.1128/jvi.64.3.1402-1406.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pescador R., Power M. D., Barr P. J., Steimer K. S., Stempien M. M., Brown-Shimer S. L., Gee W. W., Renard A., Randolph A., Levy J. A. Nucleotide sequence and expression of an AIDS-associated retrovirus (ARV-2). Science. 1985 Feb 1;227(4686):484–492. doi: 10.1126/science.2578227. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Bain E. S., Luciw P. A., Peterlin B. M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989 Apr;3(4):547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- Sheldon M., Ratnasabapathy R., Hernandez N. Characterization of the inducer of short transcripts, a human immunodeficiency virus type 1 transcriptional element that activates the synthesis of short RNAs. Mol Cell Biol. 1993 Feb;13(2):1251–1263. doi: 10.1128/mcb.13.2.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline C. T., Milocco L. H., Jones K. A. Two distinct nuclear transcription factors recognize loop and bulge residues of the HIV-1 TAR RNA hairpin. Genes Dev. 1991 Dec;5(12B):2508–2520. doi: 10.1101/gad.5.12b.2508. [DOI] [PubMed] [Google Scholar]
- Terwilliger E., Sodroski J. G., Rosen C. A., Haseltine W. A. Effects of mutations within the 3' orf open reading frame region of human T-cell lymphotropic virus type III (HTLV-III/LAV) on replication and cytopathogenicity. J Virol. 1986 Nov;60(2):754–760. doi: 10.1128/jvi.60.2.754-760.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Weiss A., Wiskocil R. L., Stobo J. D. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 production reflects events occurring at a pre-translational level. J Immunol. 1984 Jul;133(1):123–128. [PubMed] [Google Scholar]
- Wright C. M., Felber B. K., Paskalis H., Pavlakis G. N. Expression and characterization of the trans-activator of HTLV-III/LAV virus. Science. 1986 Nov 21;234(4779):988–992. doi: 10.1126/science.3490693. [DOI] [PubMed] [Google Scholar]
- Wu F. K., Garcia J. A., Harrich D., Gaynor R. B. Purification of the human immunodeficiency virus type 1 enhancer and TAR binding proteins EBP-1 and UBP-1. EMBO J. 1988 Jul;7(7):2117–2130. doi: 10.1002/j.1460-2075.1988.tb03051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Garcia J., Sigman D., Gaynor R. tat regulates binding of the human immunodeficiency virus trans-activating region RNA loop-binding protein TRP-185. Genes Dev. 1991 Nov;5(11):2128–2140. doi: 10.1101/gad.5.11.2128. [DOI] [PubMed] [Google Scholar]
- York-Higgins D., Cheng-Mayer C., Bauer D., Levy J. A., Dina D. Human immunodeficiency virus type 1 cellular host range, replication, and cytopathicity are linked to the envelope region of the viral genome. J Virol. 1990 Aug;64(8):4016–4020. doi: 10.1128/jvi.64.8.4016-4020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack J. A., Arrigo S. J., Weitsman S. R., Go A. S., Haislip A., Chen I. S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990 Apr 20;61(2):213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]






