Abstract
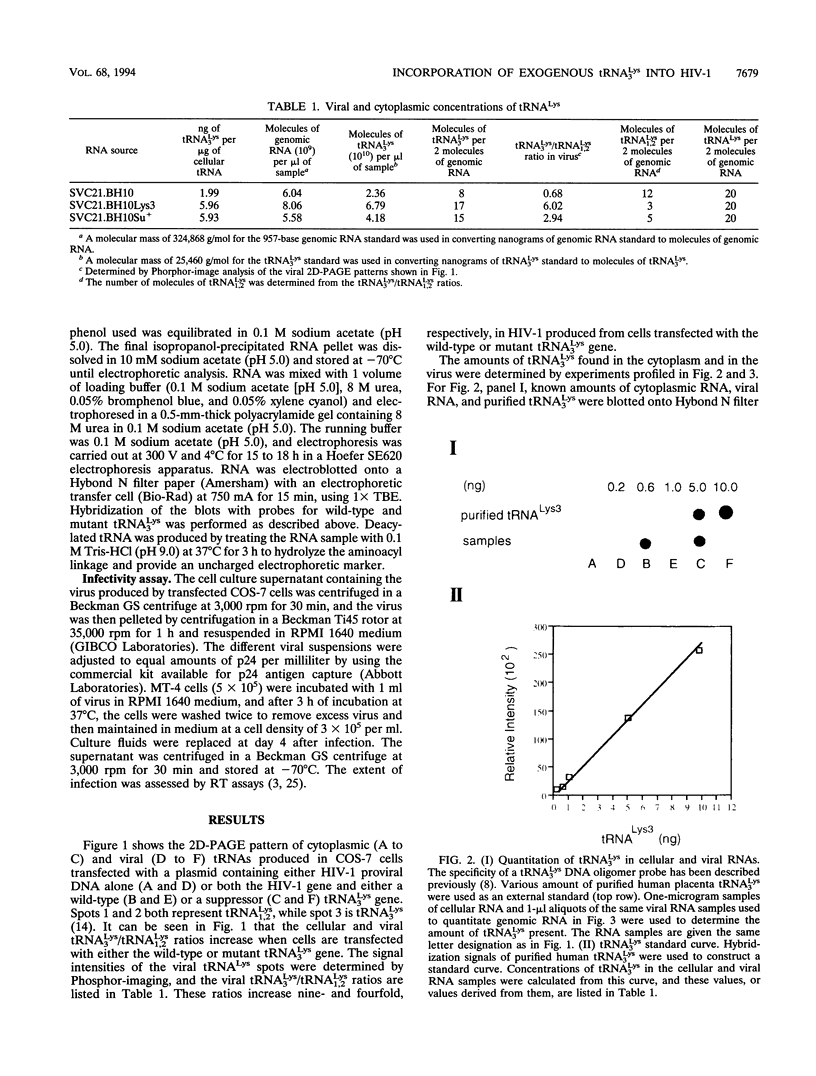
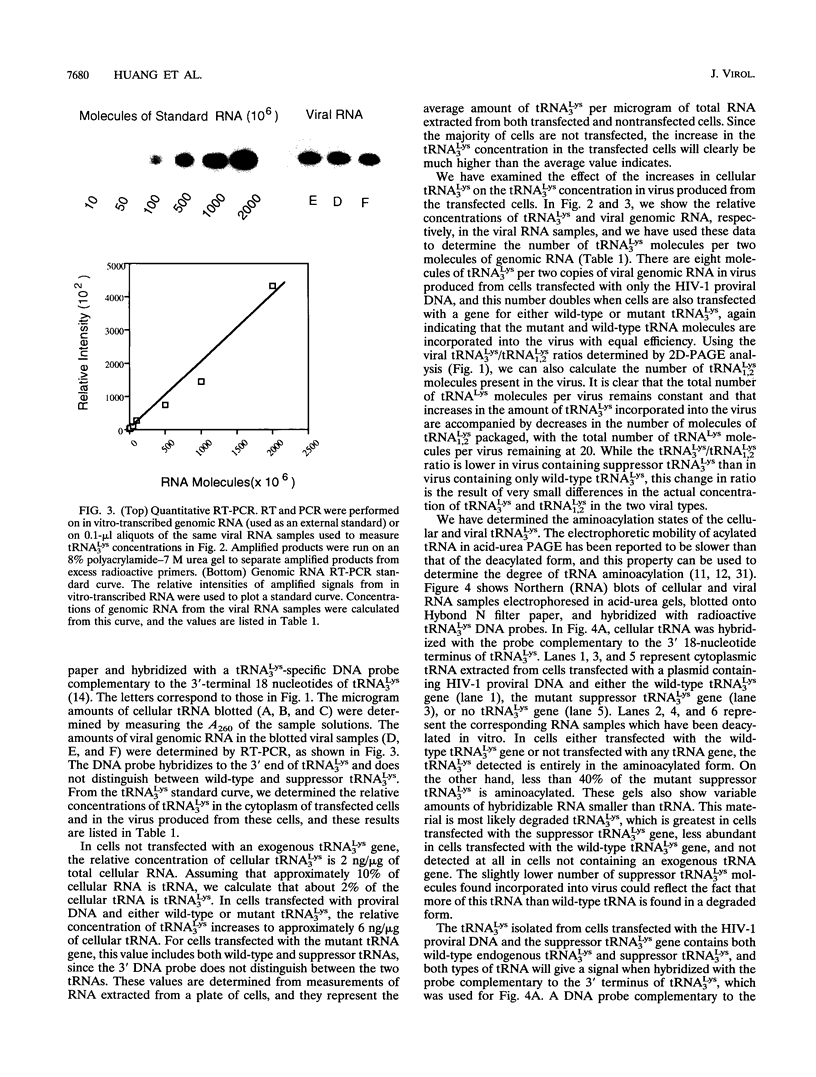
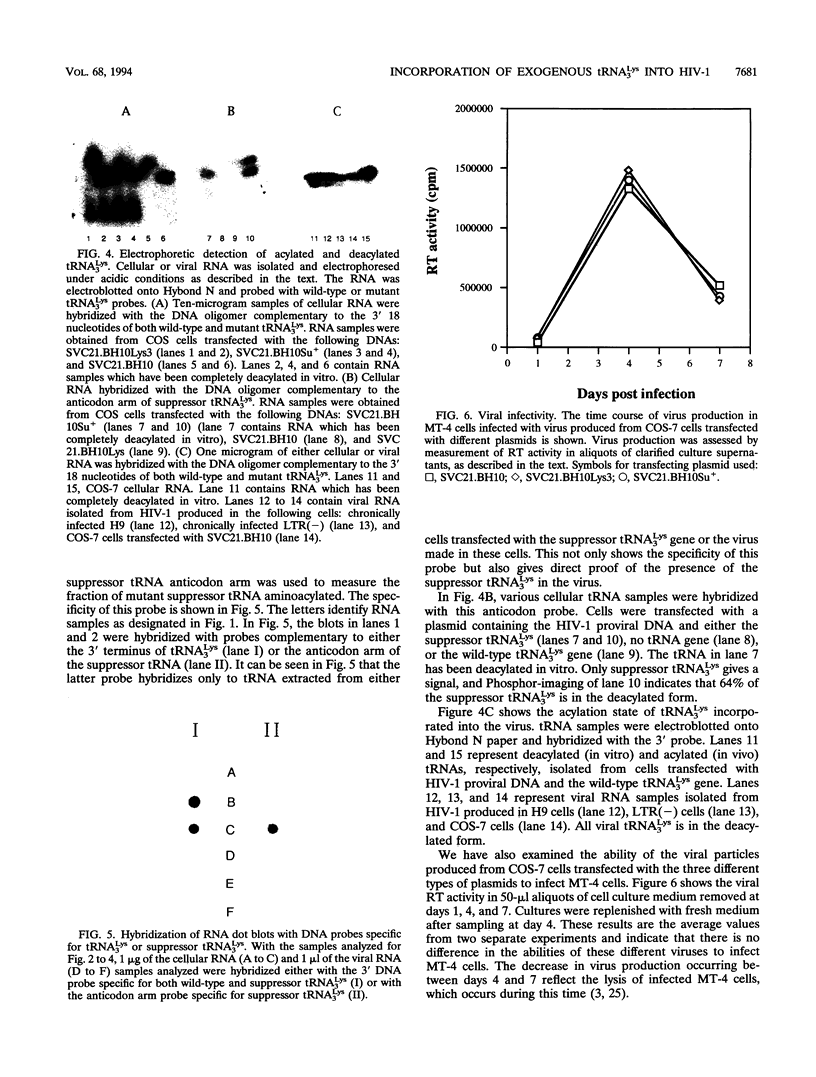
Human immunodeficiency virus (HIV) particles produced in COS-7 cells transfected with HIV type 1 (HIV-1) proviral DNA contain 8 molecules of tRNA(3Lys) per 2 molecules of genomic RNA and 12 molecules of tRNA1,2Lys per 2 molecules of genomic RNA. When COS-7 cells are transfected with a plasmid containing both HIV-1 proviral DNA and a human tRNA3Lys gene, there is a large increase in the amount of cytoplasmic tRNA3Lys per microgram of total cellular RNA, and the tRNA3Lys content in the virus increases from 8 to 17 molecules per 2 molecules of genomic RNA. However, the total number of tRNALys molecules per 2 molecules of genomic RNA remains constant at 20; i.e., the viral tRNA1,2Lys content decreases from 12 to 3 molecules per 2 molecules of genomic RNA. All detectable tRNA3Lys is aminoacylated in the cytoplasm of infected cells and deacylated in the virus. When COS-7 cells are transfected with a plasmid containing both HIV-1 proviral DNA and a mutant amber suppressor tRNA3Lys gene (in which the anticodon is changed from TTT to CTA), there is also a large increase in the relative concentration of cytoplasmic tRNA3Lys, and the tRNA3Lys content in the virus increases from 8 to 15 molecules per 2 molecules of genomic RNA, with a decrease in viral tRNA1,2Lys from 12 to 5 molecules per 2 molecules of genomic RNA. Thus, the total number of molecules of tRNALys in the virion remains at 20. The alteration of the anticodon has little effect on the viral packaging of this mutant tRNA in spite of the fact that it no longer contains the modified base mcm 5s2U at position 34, and its ability to be aminoacylated is significantly impaired compared with that of wild-type tRNA3Lys. Viral particles which have incorporated either excess wild-type tRNA3Lys or mutant suppressor tRNA3Lys show no differences in viral infectivity compared with wild-type HIV-1.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barat C., Lullien V., Schatz O., Keith G., Nugeyre M. T., Grüninger-Leitch F., Barré-Sinoussi F., LeGrice S. F., Darlix J. L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989 Nov;8(11):3279–3285. doi: 10.1002/j.1460-2075.1989.tb08488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier B., Tarrago-Litvak L., Sallafranque-Andreola M. L., Robert D., Tharaud D., Fournier M., Barr P. J., Litvak S., Sarih-Cottin L. Inhibition of the p66/p51 form of human immunodeficiency virus reverse transcriptase by tRNA(Lys). Nucleic Acids Res. 1990 Feb 11;18(3):429–436. doi: 10.1093/nar/18.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulerice F., Bour S., Geleziunas R., Lvovich A., Wainberg M. A. High frequency of isolation of defective human immunodeficiency virus type 1 and heterogeneity of viral gene expression in clones of infected U-937 cells. J Virol. 1990 Apr;64(4):1745–1755. doi: 10.1128/jvi.64.4.1745-1755.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A. G., Uhlenbeck O. C. Reactions at the termini of tRNA with T4 RNA ligase. Nucleic Acids Res. 1978 Oct;5(10):3665–3677. doi: 10.1093/nar/5.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Crawford S., Goff S. P. A deletion mutation in the 5' part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985 Mar;53(3):899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faras A. J., Dibble N. A. RNA-directed DNA synthesis by the DNA polymerase of Rous sarcoma virus: structural and functional identification of 4S primer RNA in uninfected cells. Proc Natl Acad Sci U S A. 1975 Mar;72(3):859–863. doi: 10.1073/pnas.72.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada F., Peters G. G., Dahlberg J. E. The primer tRNA for Moloney murine leukemia virus DNA synthesis. Nucleotide sequence and aminoacylation of tRNAPro. J Biol Chem. 1979 Nov 10;254(21):10979–10985. [PubMed] [Google Scholar]
- Harada F., Sawyer R. C., Dahlberg J. E. A primer ribonucleic acid for initiation of in vitro Rous sarcarcoma virus deoxyribonucleic acid synthesis. J Biol Chem. 1975 May 10;250(9):3487–3497. [PubMed] [Google Scholar]
- Ho Y. S., Kan Y. W. In vivo aminoacylation of human and Xenopus suppressor tRNAs constructed by site-specific mutagenesis. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2185–2188. doi: 10.1073/pnas.84.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Norton G. P., Palese P., Dozy A. M., Kan Y. W. Expression and function of suppressor tRNA genes in mammalian cells. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 2):1033–1040. doi: 10.1101/sqb.1986.051.01.119. [DOI] [PubMed] [Google Scholar]
- Isel C., Marquet R., Keith G., Ehresmann C., Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993 Dec 5;268(34):25269–25272. [PubMed] [Google Scholar]
- Jiang M., Mak J., Ladha A., Cohen E., Klein M., Rovinski B., Kleiman L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J Virol. 1993 Jun;67(6):3246–3253. doi: 10.1128/jvi.67.6.3246-3253.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Mak J., Wainberg M. A., Parniak M. A., Cohen E., Kleiman L. Variable tRNA content in HIV-1IIIB. Biochem Biophys Res Commun. 1992 Jun 30;185(3):1005–1015. doi: 10.1016/0006-291x(92)91727-8. [DOI] [PubMed] [Google Scholar]
- Jiang M., Parniak M. A., Kleiman L. Isolation and fractionation of retroviral tRNAs. J Virol Methods. 1992 Aug;38(2):205–213. doi: 10.1016/0166-0934(92)90111-p. [DOI] [PubMed] [Google Scholar]
- Levin J. G., Seidman J. G. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J Virol. 1981 Apr;38(1):403–408. doi: 10.1128/jvi.38.1.403-408.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin J. G., Seidman J. G. Selective packaging of host tRNA's by murine leukemia virus particles does not require genomic RNA. J Virol. 1979 Jan;29(1):328–335. doi: 10.1128/jvi.29.1.328-335.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J., Jiang M., Wainberg M. A., Hammarskjöld M. L., Rekosh D., Kleiman L. Role of Pr160gag-pol in mediating the selective incorporation of tRNA(Lys) into human immunodeficiency virus type 1 particles. J Virol. 1994 Apr;68(4):2065–2072. doi: 10.1128/jvi.68.4.2065-2072.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G. G., Hu J. Reverse transcriptase as the major determinant for selective packaging of tRNA's into Avian sarcoma virus particles. J Virol. 1980 Dec;36(3):692–700. doi: 10.1128/jvi.36.3.692-700.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Glover C. tRNA's and priming of RNA-directed DNA synthesis in mouse mammary tumor virus. J Virol. 1980 Jul;35(1):31–40. doi: 10.1128/jvi.35.1.31-40.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters G., Harada F., Dahlberg J. E., Panet A., Haseltine W. A., Baltimore D. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J Virol. 1977 Mar;21(3):1031–1041. doi: 10.1128/jvi.21.3.1031-1041.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raba M., Limburg K., Burghagen M., Katze J. R., Simsek M., Heckman J. E., Rajbhandary U. L., Gross H. J. Nucleotide sequence of three isoaccepting lysine tRNAs from rabbit liver and SV40-transformed mouse fibroblasts. Eur J Biochem. 1979 Jun;97(1):305–318. doi: 10.1111/j.1432-1033.1979.tb13115.x. [DOI] [PubMed] [Google Scholar]
- Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985 Jan 24;313(6000):277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- Rooke R., Tremblay M., Wainberg M. A. AZT (zidovudine) may act postintegrationally to inhibit generation of HIV-1 progeny virus in chronically infected cells. Virology. 1990 May;176(1):205–215. doi: 10.1016/0042-6822(90)90245-m. [DOI] [PubMed] [Google Scholar]
- Sarih-Cottin L., Bordier B., Musier-Forsyth K., Andreola M. L., Barr P. J., Litvak S. Preferential interaction of human immunodeficiency virus reverse transcriptase with two regions of primer tRNA(Lys) as evidenced by footprinting studies and inhibition with synthetic oligoribonucleotides. J Mol Biol. 1992 Jul 5;226(1):1–6. doi: 10.1016/0022-2836(92)90117-3. [DOI] [PubMed] [Google Scholar]
- Sawyer R. C., Dahlberg J. E. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J Virol. 1973 Dec;12(6):1226–1237. doi: 10.1128/jvi.12.6.1226-1237.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M. An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes. Biochim Biophys Acta. 1977 Mar 21;473(1):57–71. doi: 10.1016/0304-419x(77)90007-5. [DOI] [PubMed] [Google Scholar]
- Terwilliger E. F., Cohen E. A., Lu Y. C., Sodroski J. G., Haseltine W. A. Functional role of human immunodeficiency virus type 1 vpu. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5163–5167. doi: 10.1073/pnas.86.13.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Varshney U., Lee C. P., RajBhandary U. L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991 Dec 25;266(36):24712–24718. [PubMed] [Google Scholar]
- Waters L. C. Lysine tRNA is the predominant tRNA in murine mammary tumor virus. Biochem Biophys Res Commun. 1978 Apr 14;81(3):822–827. doi: 10.1016/0006-291x(78)91425-0. [DOI] [PubMed] [Google Scholar]
- Waters L. C., Mullin B. C., Ho T., Yang W. K. Ability of tryptophan tRNA to hybridize with 35S RNA of avian myeloblastosis virus and to prime reverse transcription in vitro. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2155–2159. doi: 10.1073/pnas.72.6.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L. C., Mullin B. C. Transfer RNA into RNA tumor viruses. Prog Nucleic Acid Res Mol Biol. 1977;20:131–160. doi: 10.1016/s0079-6603(08)60471-7. [DOI] [PubMed] [Google Scholar]
- Witte O. N., Baltimore D. Relationship of retrovirus polyprotein cleavages to virion maturation studied with temperature-sensitive murine leukemia virus mutants. J Virol. 1978 Jun;26(3):750–761. doi: 10.1128/jvi.26.3.750-761.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wöhrl B. M., Ehresmann B., Keith G., Le Grice S. F. Nuclease footprinting of human immunodeficiency virus reverse transcriptase/tRNA(Lys-3) complexes. J Biol Chem. 1993 Jun 25;268(18):13617–13624. [PubMed] [Google Scholar]