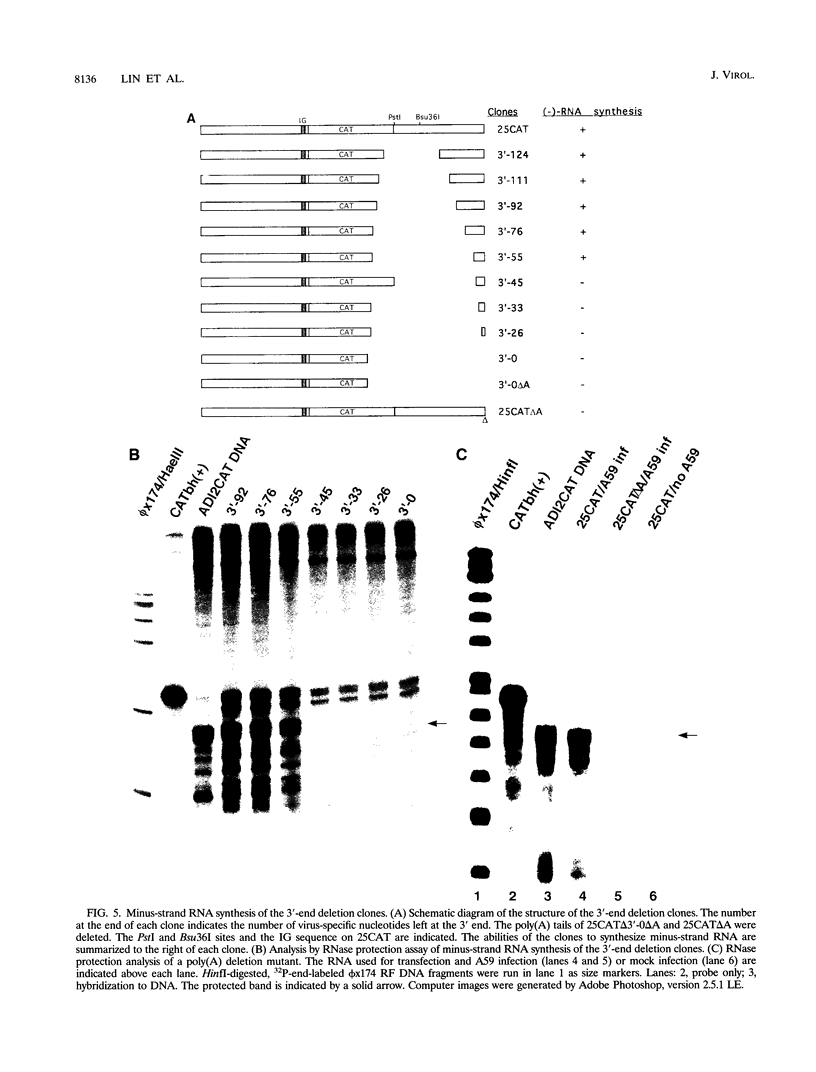
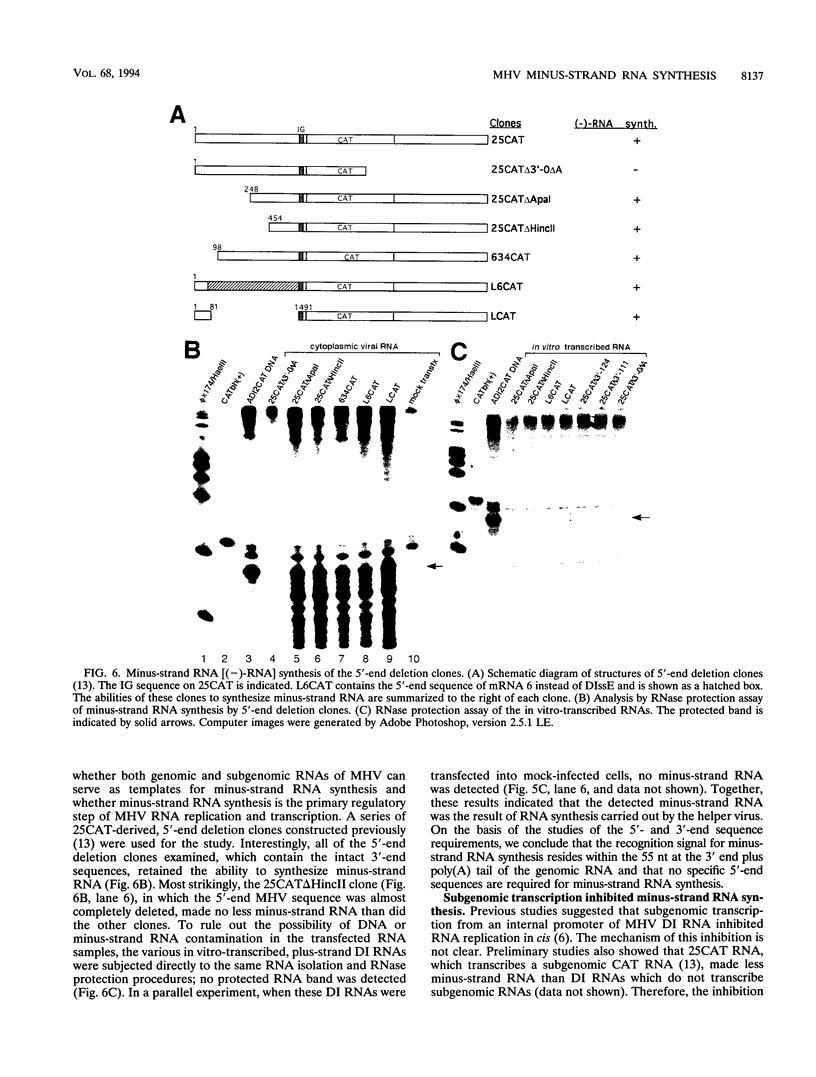
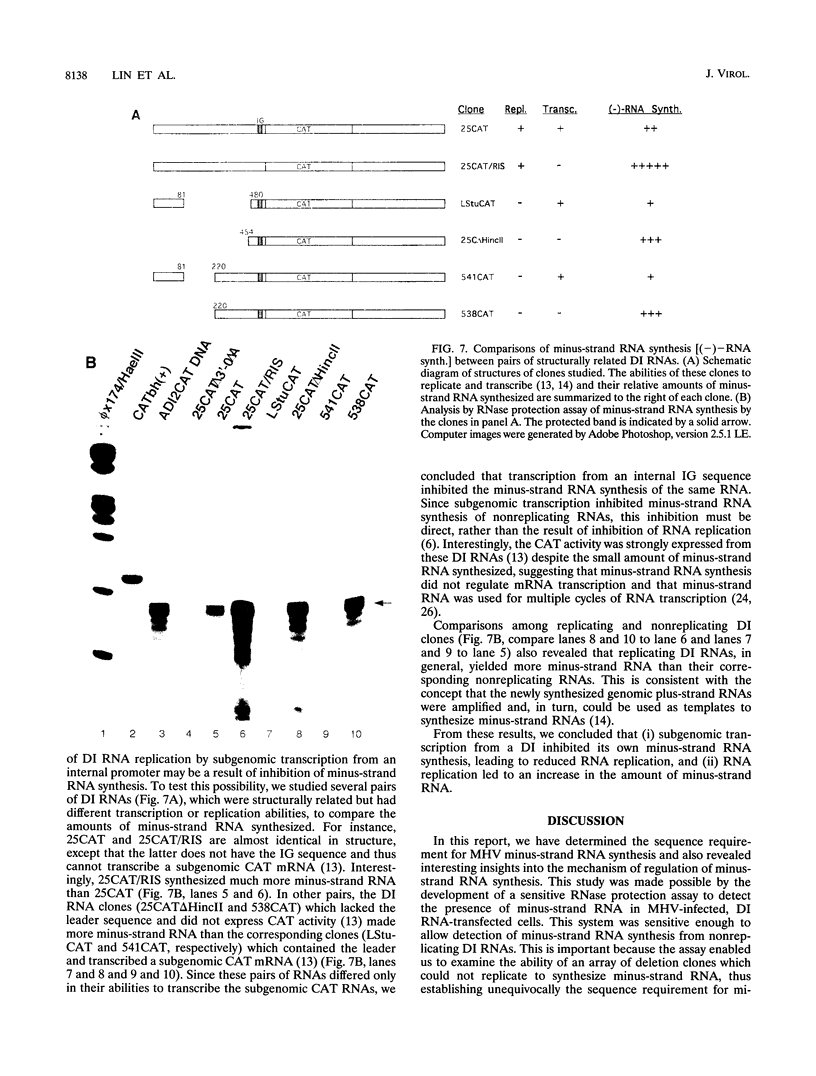
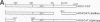Abstract
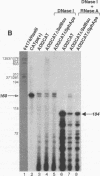
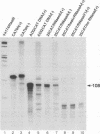
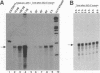
Minus-strand RNA is the first RNA species made by plus-strand RNA viruses, such as mouse hepatitis virus (MHV), and serves as a template for subsequent RNA replication and transcription. The regulation of minus-strand RNA synthesis has been difficult to study because of the paucity of minus-strand RNA. We have optimized a ribonuclease (RNase) protection assay which enabled the detection of minus-strand RNA synthesis from nonreplicating RNAs, thus clearly separating minus-strand from plus-strand RNA synthesis. We used an MHV defective interfering (DI) RNA containing a chloramphenicol acetyltransferase gene as a reporter to determine the cis-acting signal for MHV minus-strand RNA synthesis. It was found that minus-strand RNAs existed in double-stranded RNA form in the cell. By using various deletion clones, we demonstrated that the cis-acting signal for minus-strand RNA synthesis resides in the 55 nucleotides from the 3' end plus poly(A) tail of the MHV genome. This is much shorter than the 436 nucleotides previously reported for the 3'-end replication signal. No specific upstream MHV sequence was required for the initiation of minus-strand RNA synthesis. This finding suggests that the requirement for minus-strand RNA synthesis is much less stringent than that for genomic and subgenomic plus-strand RNA synthesis and that some of the minus-strand RNAs made may not be functional since they may lack the recognition signals for RNA replication or transcription. We further showed that the DI clones which actively transcribed a subgenomic mRNA from an internal intergenic sequence synthesized much less minus-strand RNA than those clones which did not transcribe subgenomic mRNAs, indicating that minus-strand RNA synthesis was inhibited by transcription from an internal promoter of the same DI RNA. This result also suggests that the regulation of the quantities of subgenomic mRNAs is not at the point of minus-strand RNA synthesis but rather at plus-strand RNA synthesis. Furthermore, the finding that the leader sequence was not required for minus-strand RNA synthesis suggests that the leader RNA regulates mRNA transcription during plus-strand RNA synthesis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dreher T. W., Hall T. C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988 May 5;201(1):31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- Furuya T., Lai M. M. Three different cellular proteins bind to complementary sites on the 5'-end-positive and 3'-end-negative strands of mouse hepatitis virus RNA. J Virol. 1993 Dec;67(12):7215–7222. doi: 10.1128/jvi.67.12.7215-7222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44(3):298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Hofmann M. A., Brian D. A. The 5' end of coronavirus minus-strand RNAs contains a short poly(U) tract. J Virol. 1991 Nov;65(11):6331–6333. doi: 10.1128/jvi.65.11.6331-6333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M. A., Sethna P. B., Brian D. A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990 Sep;64(9):4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y. S., Makino S. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J Virol. 1992 Jun;66(6):3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. N., Jeong Y. S., Makino S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993 Nov;197(1):53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Stohlman S. A. RNA of mouse hepatitis virus. J Virol. 1978 May;26(2):236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau E. T., Kong R. Y., Cheah K. S. A critical assessment of the RNAse protection assay as a means of determining exon sizes. Anal Biochem. 1993 Mar;209(2):360–366. doi: 10.1006/abio.1993.1135. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm J. A., Rice C. M. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J Virol. 1993 Apr;67(4):1905–1915. doi: 10.1128/jvi.67.4.1905-1915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C. L., Lai M. M. Requirement of the 5'-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J Virol. 1994 Aug;68(8):4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. J., Lai M. M. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontiguous sequence for replication. J Virol. 1993 Oct;67(10):6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANAKER R. A., PICZAK C. V., MILLER A. A., STANTON M. F. A hepatitis virus complicating studies with mouse leukemia. J Natl Cancer Inst. 1961 Jul;27:29–51. [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J. K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991 Nov;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Soe L. H., Baker S. C., Lai M. M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988 Oct;166(2):550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P. S., Koetzner C. A., Kerr C. A., Heo Y. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J Virol. 1994 Jan;68(1):328–337. doi: 10.1128/jvi.68.1.328-337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak J. E., Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991 Jun;65(6):3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Ries D., Bolger E., Chang L. J., Stoltzfus C. M. MHV nucleocapsid synthesis in the presence of cycloheximide and accumulation of negative strand MHV RNA. Virus Res. 1986 Dec;6(3):261–272. doi: 10.1016/0168-1702(86)90074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus minus-strand RNA synthesis and effect of cycloheximide on coronavirus RNA synthesis. J Virol. 1986 Jan;57(1):328–334. doi: 10.1128/jvi.57.1.328-334.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S. G., Sawicki D. L. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J Virol. 1990 Mar;64(3):1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hofmann M. A., Brian D. A. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J Virol. 1991 Jan;65(1):320–325. doi: 10.1128/jvi.65.1.320-325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna P. B., Hung S. L., Brian D. A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liao C. L., Lai M. M. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J Virol. 1994 Aug;68(8):4738–4746. doi: 10.1128/jvi.68.8.4738-4746.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Bredenbeek P. J., Spaan W. J. A domain at the 3' end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991 Jun;65(6):3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., de Groot R. J., Spaan W. J. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J Virol. 1994 Jun;68(6):3656–3666. doi: 10.1128/jvi.68.6.3656-3666.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]