Abstract
The alternative bacterial σN RNA polymerase holoenzyme binds promoters as a transcriptionally inactive complex that is activated by enhancer-binding proteins. Little is known about how sigma factors respond to their ligands or how the responses lead to transcription. To examine the liganded state of σN, the assembly of end-labeled Klebsiella pneumoniae σN into holoenzyme, closed promoter complexes, and initiated transcription complexes was analyzed by enzymatic protein footprinting. V8 protease-sensitive sites in free σN were identified in the acidic region II and bordering or within the minimal DNA binding domain. Interaction with core RNA polymerase prevented cleavage at noncontiguous sites in region II and at some DNA binding domain sites, probably resulting from conformational changes. Formation of closed complexes resulted in further protections within the DNA binding domain, suggesting close contact to promoter DNA. Interestingly, residue E36 becomes sensitive to proteolysis in initiated transcription complexes, indicating a conformational change in holoenzyme during initiation. Residue E36 is located adjacent to an element involved in nucleating strand separation and in inhibiting polymerase activity in the absence of activation. The sensitivity of E36 may reflect one or both of these functions. Changing patterns of protease sensitivity strongly indicate that σN can adjust conformation upon interaction with ligands, a property likely important in the dynamics of the protein during transcription initiation.
Two distinct classes of prokaryotic σ factors are known based on sequence comparison and functional differences. One class, typified by the major vegetative sigma in Escherichia coli, σ70, binds promoter elements located at −10 and −35 with respect to the initiation site, and generally binds in a transcription competent state (1). The second class, related to the E. coli σ54 (σN), activates a variety of genes in response to specific environmental stimuli (2, 3). σN confers RNA polymerase holoenzyme with properties that distinguish it from σ70 containing holoenzyme. First, the σN holoenzyme binds to distinct promoters having conserved elements at −12 and −24 (4). More importantly, σN holoenzyme binds promoters in a transcriptionally inactive state, forming stable closed complexes (4–6). Isomerization of the closed complex to transcriptionally competent open complexes requires an activator protein bound to a DNA sequence with enhancer-like properties (7, 8) and nucleoside triphosphate hydrolysis by the activator protein (5, 9, 10). Activation appears to involve direct contact between activator and the holoenzyme (11–13).
The function of several regions of σN have been assessed through mutagenesis. The conserved N-terminal 50 amino acids (region I) contain an excess of glutamine residues and characteristic regularly repeated leucines and are required for response to activator proteins (14–17). Interestingly, certain mutations in this domain result in low-level activator-independent transcription (18, 19). The next 50 or so amino acids (region II) are not conserved in sequence or in number, but are generally acidic and for E. coli σN contribute to DNA melting (14, 20). The carboxyl-terminal two-thirds of the protein (region III) is highly conserved and performs at least three functions. First, residues 70–215 interact with core RNA polymerase (15, 21, 22). Next, residues 329–477 comprise the primary DNA binding determinant (15, 23, 24) and include a predicted helix–turn–helix motif (25) and the rpoN box, a characteristic element found in all σN proteins. Mutations in either of these sequence elements can alter DNA binding (26, 27). Finally, residues 180–306 contribute to DNA binding activity by enhancing binding of the minimal DNA binding domain (28).
Partial proteolysis, or “protease footprinting,” has been used to detect protein–protein, protein–DNA, and protein–ligand interactions (29–33). Both contacts between protein and ligand (30, 32) and conformational changes in the proteins (34, 35) have been inferred. We have used partial proteolysis of 32P end-labeled σN [labeled through introduction of a heart muscle protein kinase (HMK) site, −HMK] as a means of examining the interactions of σ with core, DNA, and in open complexes. Our results indicate that surface-exposed regions of σN undergo significant conformational changes as a result of interaction with core, and during the transition from closed to open complexes. In addition, amino acid residues in direct contact or close proximity to DNA have been identified. The changes in protease sensitivity resulting from these interactions demonstrate that localized conformational changes in σN accompany each step along the initiation pathway. Such changes may reflect movements that are necessary for the normal function of σN.
MATERIALS AND METHODS
Overexpression and Purification of σN-HMK.
The plasmid pHMK3′ encodes the Klebsiella pneumoniae σN protein modified by precise addition of the five-residue tag RRASV at the C terminus, which allows C-terminal specific labeling with γ32P-ATP using HMK. It was constructed by PCR amplification of the rpoN gene in pMM70 (36) by using an upstream universal primer, 5′-GTTTTCCCAGTCACGAC and a downstream primer, 5′-ATAGTGAAGCTTCAAACAGATGCACGACGAACCAGCTGCTTGCGCTG, encoding the sequence RRASV (in italics) followed by a stop codon and a HindIII restriction site (underline). A 150-bp SacII–HindIII fragment from the PCR product was substituted for the equivalent wild-type fragment from pWVC93025 (37).
Expression of σN-HMK was induced in E. coli strain 71–18 by addition of isopropyl β-d-thiogalactoside to 1 mM for 2 hr at 30°C. Cells from 2 liters of culture were lysed for 30 min in 15 ml of 50 mM Tris, pH 8.0/10 mM EDTA/5% glycerol/50 mM NaCl/1 mM DTT/1 mM phenylmethylsulfonyl fluoride/0.2% sodium deoycholate/200 μg/ml lysozyme, followed by sonication to reduce viscosity of the extract. Streptomycin sulfate was added to 0.2% final concentration, and supernatant was recovered after centrifugation at 40,000 × g for 30 min. Ammonium sulfate was added to 70% saturation, and the precipitate was recovered after centrifugation. The pellet was resuspended in 5 ml of TGED (20 mM Tris, pH 8.0/5% glycerol/0.1 mM EDTA/1 mM DTT), dialyzed overnight at 4°C against TGED. Dialyzed extract was purified as described for wild-type σN with minor modifications (24). Briefly, chromatography was performed over DEAE-Sepharose in imidazole buffer and eluted with a gradient of NaCl (σN-HMK elutes at 300–350 mM NaCl). Further purification was with heparin agarose in TGED (σN-HMK elutes at 250 mM NaCl), and Resource Q anion exchange resin in TGED (σN-HMK elutes at 350 mM NaCl). After purification the sample was dialyzed against TGED + 50% glycerol/50 mM NaCl and stored at −70°C (long-term storage) or −20°C (short-term storage).
Labeling with γ32P-ATP.
Two micrograms of σN-HMK was labeled by using 50 μCi (5,000 Ci/mmol) γ 32P-ATP and 0.5 unit HMK (Sigma) for 20 min on ice in 20 mM Tris, pH 7.5/100 mM NaCl/12 mM MgCl2/4 mM DTT in a final volume of 10 μl. Under these conditions wild-type σN is not detectably labeled. Typically, approximately 10% of the tagged protein was labeled. The labeling reaction was used directly, or in some cases unincorporated ATP was removed by repeated concentration in Nanosep microconcentrators. Aliquots of labeled protein were stored at −70°C and used only once after thawing.
Purification of PspF.
Strain K1629 (gift of G. Jovanovic and P. Model, The Rockefeller University, New York) was used for overexpression of the E. coli activator protein ΔPspF (38). This activator naturally lacks an N-terminal regulatory domain and has a disrupted DNA binding domain. This activator has the advantages that it is soluble when overexpressed, and it is constitutively active. Extracts of soluble protein were prepared as for σN-HMK. The N-terminally hexahistidine-tagged protein then was purified by Ni-affinity chromatography using HiTrap chelating resin (Pharmacia) in 20 mM Tris, pH 7.0/500 mM NaCl and eluted with an imidazole gradient of 20–500 mM. PspF elutes at around 350 mM imidazole. After dialysis against TGED, the protein was purified further by anion exchange chromatography using a Resource Q column equilibrated in TGED and eluted with a 50–500 mM NaCl gradient. The PspF peak was at 300 mM NaCl. Aliquots of the purified protein in TGED/50 mM NaCl were stored at −70°C.
Assembly of Complexes.
Holoenzyme was formed in 9-μl reactions by mixing approximately 10 ng σN-HMK with a 1.5- to 2-fold molar excess of either K. pneumoniae or E. coli (Epicentre Technologies, Madison WI) core polymerase in TGED/50 mM NaCl/5 mM MgCl2 at 25°C for 5 min. An equal amount (mass) of BSA was added to reactions lacking core polymerase. Closed complexes were formed in an identical manner except that after holoenzyme assembly, 1 μg of a 650-bp DNA fragment containing the Rhizobium meliloti nif H promoter was added, and the reactions were incubated an additional 10 min at 30°C. The promoter fragment was prepared by PCR amplification of the promoter and nifA UAS in pMB210.1 (39) using the primers as follows: 5′-AAAAATAGGCGTATCACGAGGCCCT (upstream) and 5′-TATTTCGGTTGTTCGGACACATGAA (downstream). The 650-bp product was purified by agarose gel electrophoresis, extracted from gel slices using Qiagen resin, then phenol/chloroform-extracted and ethanol-precipitated before use. Open complexes were prepared by mixing 30 ng 32P end-labeled σN-HMK, 70 ng unlabeled σN-HMK, 720 ng E. coli core RNA polymerase, and 1 μg promoter DNA in 12 μl 50 mM tris-acetate/10 mM potassium acetate/8 mM magnesium acetate/1 mM DTT/3.5% (wt/vol) polyethylene glycol 6,000–8,000, pH 8.0 (STA). After 10 min at 30°C, purified PspF and GTP were added to 4 μM and 1 mM, respectively. After an additional 10-min incubation, protease was added as needed. Reactions were stopped by addition of heparin to 100 μg/ml and 3,4 dichloroisocoumarin to 0.25 mM and chilling on ice. Open complexes were isolated after electrophoresis through 4% polyacrylamide gels in 20 mM Tris/200 mM glycine, pH 8.3 buffer. Isolated gel slices were incubated for 15 min in 15 μl 2× SDS/PAGE loading buffer before separation by SDS/PAGE.
Protease Digestions.
All digestions were performed at 25°C for 3.5 min and contained approximately 10 ng of 32P-labeled σN-HMK in TGED except for open complexes. Quantitation of the digestions by using a PhosphorImager demonstrated that cleavage of less then 10% of the σN-HMK produced a nearly identical set of products as higher levels of cleavage, indicating that the main digestion products result from a single cleavage. Addition of DNA alone, or BSA in amounts equal to that of RNA polymerase or PspF used in these experiments, had little effect on the extent of digestion so the same conditions were used in all reactions. Digestions were stopped by addition of 3× SDS/PAGE loading buffer containing 0.5 mM 3,4 dichloroisocoumarin and heating immediately to 95°C for 5 min. Products were separated by SDS/PAGE on 15% Lamelli polyacrylamide gels for fragments 12–65 kDa or on 16.5% Tris-tricine gels (40) for fragments 6–25 kDa. Cleavage sites were mapped based on the predicted sequence specificity of the proteases, comparison with migration of purified, defined truncated σ proteins, and previously sequenced chymotryptic peptides (23, 24).
RESULTS
Tagged σN-HMK.
pHMK 3′ encodes the full-length K. pneumoniae rpoN gene followed by the sequence RRASV, which allows phosphorylation by bovine HMK. The HMK-tagged protein retains activity comparable to wild-type σN in vivo and purifies as wild type. The purified tagged protein binds core RNA polymerase normally as measured in gel mobility shift assays and binds promoter DNA normally in exonuclease III and ortho-copper phenanthroline DNA footprinting assays (data not shown). Formation of heparin-resistant open complexes on labeled promoter fragments also appeared normal. Titrations of proteases were performed, and partial digestion conditions were established. The most useful proteases were found to be the V8 protease, chymotrypsin, and subtilisin, each of which produced a distinct set of bands. Cleavage of less than 10% and up to approximately 50% of σN-HMK molecules resulted in nearly identical sets of products, indicating that the major products result from primary cleavage. The sites of cleavage were identified by using three σN standards in addition to standard molecular mass markers. First, the positions of cleavage sites in regions I and II were mapped based on migration relative to purified N-terminally deleted σN proteins (24). Second, previously sequenced chymotryptic peptides beginning at residues 304 and 329 allowed identification of the V8 cleavage site at E325. Finally, urea denatured 32P-labeled σN-HMK was digested with trypsin and the sites of cleavage were determined based on the calculated molecular masses using low molecular mass standards (Sigma 17S) to generate the standard curve. Cleavage at R383, K388, and R394 was assigned in this way and provided a useful marker for estimation of the V8 cleavage sites in the DNA binding domain. Our assay provides information about a number of residues in σ, but is limited in that no cleavage sites are observed in most of region I, or between residues 135–303, suggesting the latter may form a tightly folded protein core.
Holoenzyme Formation.
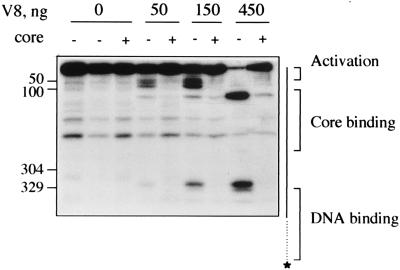
The effects of core RNA polymerase on proteolysis of σN-HMK are shown in Figs. 1 and 2. Free σN-HMK is sensitive to V8 protease at several sites; one region comprised of several sites extending from near residue 36 to residue 100 (+/− 5 residues), and further sites at residues 135 and 325. Cleavage at residue 334 is weak and is not always clearly present, suggesting it may be a secondary cleavage product. Core RNA polymerase protects two distinct regions from proteolytic cleavage, one near the N terminus from residues 36–100, and a second at residues 325/334. (Compare Fig. 1, lanes 4, 6, and 8 with lanes 5, 7, and 9). Subtilisin cleaves σN-HMK at sites overlapping the V8 sites (Fig. 2, lane 10), although because of the broad specificity of subtilisin it is not possible to specify exact residues. Core polymerase prevents subtilisin cleavage between residues 304 and 328 (Fig. 2, lanes 10 and 11). In contrast to V8 protease, subtilisin cleavage sites near residue 100 are unaffected. In the case of chymotrypsin digestions, core polymerase sometimes partially protected residue Y303 from digestion (Fig. 2, lanes 6 and 7); however, this protection was not seen in all experiments.
Figure 1.
V8 cleavage of polymerase holoenzyme. Migration of sequenced σN-HMK chymotryptic peptides ending at residues 304 and 329, and of region I (50) or region I+II deleted σ (100) is shown on the left. A diagram of σN-HMK on the right shows the locations of region I and core binding domain. A Tris-glycine SDS buffer system was used. Lanes 1-3, undigested σN-HMK incubated with no protein, 70 ng BSA, or 70 ng K. pneumoniae core RNA polymerase, respectively. Lanes 4, 6, and 8, σN-HMK + BSA, digested with V8 protease. Lanes 5, 7, and 9, σN-HMK holoenzyme digested with V8 protease.
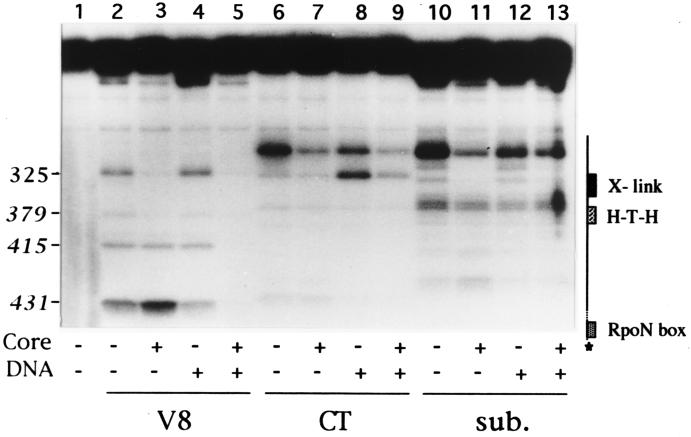
Figure 2.
Proteolytic cleavage of closed complexes. Migration of σN-HMK chymotryptic peptides ending at residues 304 and 329, and probable ends of major V8 products (in italics) is indicated on the left. Sites of V8 cleavage were mapped based on calculated molecular weights using Sigma 17S markers as standards, and comparison with tryptic and V8 digests of urea denatured σN-HMK. The diagram on the right shows the positions of previously defined DNA binding domain elements. Lane 1, undigested σN-HMK. Lanes 2–5, V8 digestions (50 ng). Lanes 6–9, chymotrypsin digestions (12.5 ng). Lanes 10–13, subtilisin digestions (1 ng). Core is E. coli core RNA polymerase. DNA is 1 μg R. meliloti nifH promoter fragment extending from −600 to +40. Proteolytic fragments were resolved on a 16.5% polyacrylamide gel using a Tris-tricine-SDS buffer system.
Products from full-length σN down to approximately 12 kDa (residues 1–350) are resolved in the Tris-glycine electrophoresis buffer used in the V8 digestions described above. A Tris-tricine buffer system was used to resolve products with molecular masses down to approximately 6 kDa (residues 300–430). These experiments revealed several protease sensitive sites located between residues 350 and 430 (Fig. 2, lanes 2, 6, and 10). Among these sites, only one, the V8 protease site at residue E378, is protected by core (Fig. 2, lane 3). Overall, these results demonstrate that specific protease-sensitive sites in σN-HMK are protected in the holoenzyme complex. The protected sites lie outside the minimal core binding domain but within regions involved in response to activators (residues 36–50), DNA melting (residues 50–100), and adjacent to or within the DNA binding domain (residues 304–327 and 378). Because these changes lie outside the minimal core binding domain, they most likely are due to conformational changes in σN-HMK that occur upon interaction with core.
Closed Complex-Specific Protection.
A DNA fragment containing the R. meliloti nifH promoter was added to holoenzyme complexes to form closed complexes, and these were subjected to proteolysis. The resulting patterns of cleavage sites with all three proteases in the amino terminal 380 residues were similar to the corresponding holoenzyme patterns (Fig. 2, lanes 3 and 5, 7 and 9, 11 and 13, and data not shown). However beyond residue 380 two V8 cleavage sites clearly are protected from digestion in the closed complex. These sites, most probably located at residues E410/414 and E431, lie within the minimal DNA binding domain of σN, between the predicted helix–turn–helix motif and the rpoN box. Both of these elements have been implicated in promoter recognition (26, 27, 37, 41, 42), so it is plausible that protection of E410/414 and E431 results from their proximity to DNA. No reproducible differences were seen between holoenzyme and closed complexes with chymotrypsin or subtilisin digestion. Free wild-type σN binds DNA specifically (43), so it is predicted that E410/414 and E431 would be protected in σ-DNA complexes. We looked for such an interaction but were unable to detect one, perhaps due to low affinity or transient σ-DNA interactions (Fig. 2, lane 4). A moderate increase in sensitivity to cleavage with chymotrypsin at residue W328 was observed consistently, however (Fig. 2, lane 8), suggesting that conformational changes outside the DNA binding domain occur in response to DNA binding.
Region I Cleavage Is Enhanced in Open Complexes.
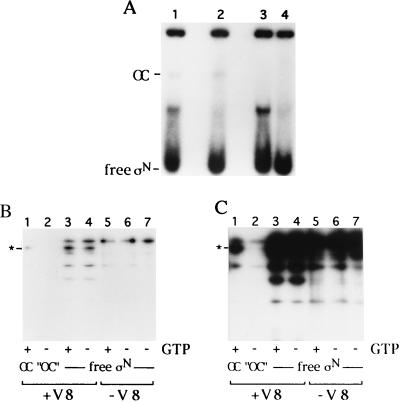
Activators of σN-holoenzyme function by transforming closed complexes into open complexes that are heparin stable in a mechanism dependent upon nucleoside triphosphate hydrolysis. We have used a derivative of the activator protein PspF from E. coli to generate open complexes. Although this derivative lacks a DNA binding domain, it promotes DNA melting at high concentrations (500 μM–4 mM). Whereas end-labeled σN-HMK was incorporated into holoenzyme and closed complexes quantitatively, only a fraction was incorporated into open complexes on the linear template, as observed by others (5, 44, 45). Therefore, open complexes were purified by preparative native gel electrophoresis after proteolysis and before analysis by SDS/PAGE. The results of one such experiment are shown in Fig. 3 A and B. The complexes examined were initiated complexes because the nucleotide used for the activator, GTP, also allows transcription of the first three nucleotides of the nifH mRNA used in these experiments. GTP was used to enhance the yield of open complexes. Fig. 3A shows the autoradiograph of a native gel used for open complex isolation. An open complex band can be seen in reactions containing GTP, but is absent from reactions minus GTP (Fig. 3A, lanes 1 and 2). Digestion with V8 protease results in little change in the open complex band but does result in a GTP-independent appearance of a faster migrating band. Several bands were isolated from this gel and analyzed further by SDS/PAGE (Fig. 3B). Cleaved free σN-HMK isolated from the gels exhibits a pattern of products similar to that of free σ loaded directly onto denaturing gels (Fig. 3B, lanes 4 and 5). Interestingly, among the V8 cleavage sites in regions I and II that we demonstrated are protected in holoenzyme and closed complexes a single site within region I becomes sensitive to proteolysis in open complexes (Fig. 3B, lane 7). This site, whose location is most likely E36, lies adjacent to a sequence element shown by mutagenesis to be required for DNA strand separation and to inhibit the transition from closed to open complexes in the absence of activation (19, 46, 47). We propose that protease sensitivity at residue E36 is a reflection of a conformational change that occurs in this domain as a result of activation.
Figure 3.
Proteolysis of open complexes. (A) Isolation of open complex on 4% native polyacrylamide gel. Each reaction contained 100 nM σN-HMK, 100 nM E. coli core RNA polymerase, 150 nM nif H DNA, and 4 μM PspF. Reactions in lanes 1 and 2 contained GTP. Reactions in lanes 3 and 4 lack GTP. Bands corresponding to V8 digested (lanes 1 and 3) and undigested (lanes 2 and 4) open complexes and free σN-HMK were cut out of the gel, as was a portion of the gel in the −GTP lane with the same migration as the open complex as a control for background. (B) SDS/PAGE analysis of isolated bands from native gel. Lane 1, free σN-HMK, not gel-purified. Lanes 2 and 3, free σN-HMK, undigested, isolated from lanes 2 and 4 of A. Lanes 4 and 5, free σN-HMK, digested with V8, isolated from lanes 1 and 3 of A. Lanes 6 and 7, open complex, or “mock open complex,” digested with V8, isolated from lanes 1 and 3. (C) Longer exposure of B.
DISCUSSION
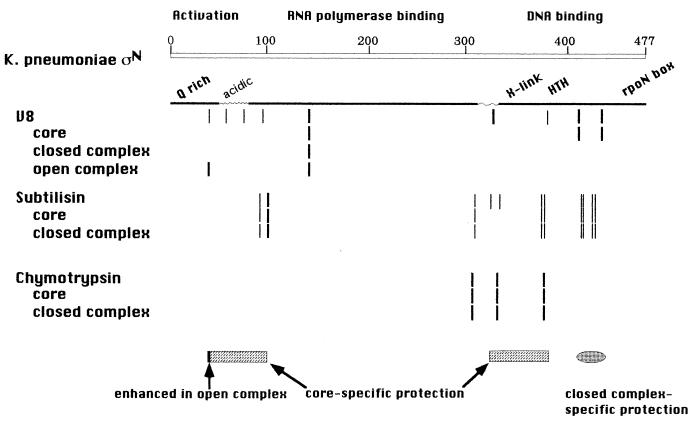
We have shown that σN undergoes specific alterations in protease sensitivity at different steps on the pathway leading from free σN to an engaged transcription complex. The patterns of protease sensitivity are summarized in Fig. 4. The residues protected when holoenzyme is formed most probably are due to conformational changes rather than specific σ-core polymerase interactions for two reasons. First, these changes occur mainly outside the minimal core binding domain of σN, which extends from 70–215 (M.-T. Gallegos, personal communication). Second, the protected region II is not conserved among σN proteins and therefore is unlikely to make specific contacts with core. It is possible that the protection is due to an interaction of region II with core. Because holoenzyme binds promoters differently both quantitatively and qualitatively from free σN (46, 47) it is plausible that the predicted conformational changes result in altered DNA binding activity. In this regard the protection of E378 from V8 cleavage in the holoenzyme is particularly interesting, as it lies in a region involved specifically in DNA binding and intolerant to mutagenesis (15, 24, 26, 27). In addition, region I influences DNA binding around the −12 element (15–17, 47), suggesting that region I conformational changes could modify σN-DNA interactions. A structure in which region I is in close proximity to the DNA binding domain could account for both protection of noncontiguous sequences in sigma by core and the influence of region I on DNA binding activity.
Figure 4.
Summary map of protease-sensitive sites. Vertical lines represent digestion sites. Rectangles and ovals at the bottom indicate the region protected by core polymerase and in closed complexes, and the dark vertical line indicates position of the sensitive site in initiated complexes.
The closed complex-specific protections also can be interpreted as conformational alterations, but because protected residues E410/414 and E431 lie within the DNA-binding domain, it is also possible that protection results from close contact with promoter DNA. Both a predicted helix–turn–helix motif at residues 367–386 and the rpoN box at residues 454–463 have been implicated in DNA binding activity (26, 41, 42). Additionally, deletion mutants spanning the C-terminal one-third of σN and random mutagenesis of residues 307–392 suggests that DNA binding determinants are spread throughout this region (15, 27). Interestingly, in an analysis of amino acid-DNA interactions in crystal structures, it appears that glutamate residues have a distinct preference for C residues (48). Given that the σN binding site consensus sequence is GC-rich, CTGGCGN5TTCG, it may be that residues 410/414 and 431 make specific contacts with promoter C residues.
A striking result of these studies was the enhanced cleavage seen at residue E36 of region I in initiated complexes compared with closed complexes. Region I is required for the response to activator proteins and has been proposed to perform two functions. The first is to promote initiation in the presence of an activator, and the second is to maintain a closed complex in the absence of activator (19, 47). Mutations adjacent to E36 can affect either activity. Thus enhanced cleavage at E36 may be a reflection of either or both of these functions. The increased cleavage we observe could, in principle, be due to changes in σN itself or to the core RNA polymerase. Considering the essential role played by region I in σN function, we prefer a model in which conformational change around E36 actively promotes open complex formation.
Acknowledgments
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: HMK, heart muscle kinase; TGED, 20 mM Tris, pH 8.0/5% glycerol/0.1 mM EDTA/1 mM DTT.
References
- 1.Lonetto M, Gribskov M, Gross C A. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kustu S, Santero E, Keener J, Popham D, Weiss D. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merrick M J. Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 4.Morett E, Buck M. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 5.Popham D L, Szeto D, Keener J, Kustu S. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 6.Sasse-Dwight S, Gralla J D. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morett E, Cannon W, Buck M. Nucleic Acids Res. 1988;16:11469–11488. doi: 10.1093/nar/16.24.11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reitzer L J, Magasanik B. Cell. 1986;45:785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- 9.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 10.Austin S, Dixon R. EMBO J. 1991;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoover T R, Santero E, Porter S, Kustu S. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 12.Lee J H, Hoover T R. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su W, Porter S, Kustu S, Echols H. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasse-Dwight S, Gralla J D. Cell. 1990;62:945–954. doi: 10.1016/0092-8674(90)90269-k. [DOI] [PubMed] [Google Scholar]
- 15.Wong C, Tintut Y, Gralla J D. J Mol Biol. 1994;236:81–90. doi: 10.1006/jmbi.1994.1120. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh M, Gralla J D. J Mol Biol. 1994;239:15–24. doi: 10.1006/jmbi.1994.1347. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh M, Tintut Y, Gralla J D. J Biol Chem. 1994;269:373–378. [PubMed] [Google Scholar]
- 18.Syed A, Gralla J D. Mol Microbiol. 1997;23:987–995. doi: 10.1046/j.1365-2958.1997.2851651.x. [DOI] [PubMed] [Google Scholar]
- 19.Wang J T, Syed A, Hsieh M, Gralla J D. Science. 1995;270:992–994. doi: 10.1126/science.270.5238.992. [DOI] [PubMed] [Google Scholar]
- 20.Wong C, Gralla J D. J Biol Chem. 1992;267:24762–24768. [PubMed] [Google Scholar]
- 21.Tintut Y, Wong C, Jiang Y, Hsieh M, Gralla J D. Proc Natl Acad Sci USA. 1994;91:2120–2124. doi: 10.1073/pnas.91.6.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tintut Y, Gralla J D. J Bacteriol. 1995;177:5818–5825. doi: 10.1128/jb.177.20.5818-5825.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cannon W, Claverie-Martin F, Austin S, Buck M. Mol Microbiol. 1994;11:227–236. doi: 10.1111/j.1365-2958.1994.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 24.Cannon W, Missailidis S, Smith C, Cottier A, Austin S, Moore M, Buck M. J Mol Biol. 1995;248:781–803. doi: 10.1006/jmbi.1995.0260. [DOI] [PubMed] [Google Scholar]
- 25.Merrick M, Gibbins J, Toukdarian A. Mol Gen Genet. 1987;210:323–330. doi: 10.1007/BF00325701. [DOI] [PubMed] [Google Scholar]
- 26.Taylor M, Butler R, Chambers S, Casimiro M, Merrick M. Mol Microbiol. 1996;22:1045–1054. doi: 10.1046/j.1365-2958.1996.01547.x. [DOI] [PubMed] [Google Scholar]
- 27.Guo Y, Gralla J D. J Bacteriol. 1997;179:1239–1245. doi: 10.1128/jb.179.4.1239-1245.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cannon W V, Chaney M K, Wang X-Y, Buck M. Proc Natl Acad Sci USA. 1997;94:5006–5011. doi: 10.1073/pnas.94.10.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greiner D P, Hughes K A, Gunasekera A H, Meares C F. Proc Natl Acad Sci USA. 1996;93:71–75. doi: 10.1073/pnas.93.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogenhagen D F. Mol Cell Biol. 1993;13:5149–5158. doi: 10.1128/mcb.13.9.5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heyduk T, Heyduk E, Severinov K, Tang H, Ebright R H. Proc Natl Acad Sci USA. 1996;93:10162–10166. doi: 10.1073/pnas.93.19.10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hori R, Sung P, Carey M. Proc Natl Acad Sci USA. 1995;92:6047–6051. doi: 10.1073/pnas.92.13.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyduk E, Heyduk T. Biochemistry. 1994;33:9643–9650. doi: 10.1021/bi00198a033. [DOI] [PubMed] [Google Scholar]
- 34.Kenney L J, Bauer M D, Silhavy T J. Proc Natl Acad Sci USA. 1995;92:8866–8870. doi: 10.1073/pnas.92.19.8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts S G E, Green M R. Nature (London) 1994;371:717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- 36.Merrick M J, Gibbins J R. Nucleic Acids Res. 1985;13:7607–7620. doi: 10.1093/nar/13.21.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cannon W, Austin S, Moore M, Buck M. Nucleic Acids Res. 1995;23:351–356. doi: 10.1093/nar/23.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jovanovic G, Weiner L, Model P. J Bacteriol. 1996;178:1936–1945. doi: 10.1128/jb.178.7.1936-1945.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buck M, Cannon W. Nucleic Acids Res. 1989;17:2597–2612. doi: 10.1093/nar/17.7.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schagger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 41.Merrick M, Chambers S. J Bacteriol. 1992;174:7221–7226. doi: 10.1128/jb.174.22.7221-7226.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppard J R, Merrick M J. Mol Microbiol. 1991;5:1309–1317. doi: 10.1111/j.1365-2958.1991.tb00777.x. [DOI] [PubMed] [Google Scholar]
- 43.Buck M, Cannon W. Nature (London) 1992;358:422–424. doi: 10.1038/358422a0. [DOI] [PubMed] [Google Scholar]
- 44.Wedel A, Kustu S. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 45.Eydmann T, Soderback E, Jones T, Hill S, Austin S, Dixon R. J Bacteriol. 1995;177:1186–1195. doi: 10.1128/jb.177.5.1186-1195.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cannon W, Claverie-Martin F, Austin S, Buck M. Mol Microbiol. 1993;8:287–298. doi: 10.1111/j.1365-2958.1993.tb01573.x. [DOI] [PubMed] [Google Scholar]
- 47.Morris L, Cannon W, Claverie-Martin F, Austin S, Buck M. J Biol Chem. 1994;269:11563–11571. [PubMed] [Google Scholar]
- 48.Mandel-Gutfreund Y, Schueler O, Margalit H. J Mol Biol. 1995;253:370–382. doi: 10.1006/jmbi.1995.0559. [DOI] [PubMed] [Google Scholar]