Abstract
Cells of the budding yeast Saccharomyces cerevisiae are born carrying localized transmembrane landmark proteins that guide the subsequent establishment of a polarity axis and hence polarized growth to form a bud in the next cell cycle. In haploid cells, the relevant landmark proteins are concentrated at the site of the preceding cell division, to which they recruit Cdc24, the guanine nucleotide exchange factor for the conserved polarity regulator Cdc42. However, instead of polarizing at the division site, the new polarity axis is directed next to but not overlapping that site. Here, we show that the Cdc42 guanosine triphosphatase–activating protein (GAP) Rga1 establishes an exclusion zone at the division site that blocks subsequent polarization within that site. In the absence of localized Rga1 GAP activity, new buds do in fact form within the old division site. Thus, Cdc42 activators and GAPs establish concentric zones of action such that polarization is directed to occur adjacent to but not within the previous cell division site.
Introduction
During development and differentiation, it is sometimes important for cells to position specific structures adjacent to but not overlapping each other. For example, in epithelia, apical tight junctions are found next to more basal adherens junctions. In haploid Saccharomyces cerevisiae, new buds form next to the previous cell division site but never within it. Because yeast cell division is accompanied by deposition of specialized “bud scars” on the mother cell wall and “birth scars” on the daughter cell wall (Barton, 1950; Chant and Pringle, 1995), it could be that the scars physically preclude subsequent bud emergence at division sites. Indeed, an early hypothesis to explain reproductive aging in yeast was that bud scars would eventually cover the entire cell wall (Mortimer and Johnson, 1959). However, even if cell wall scars impede budding, how would cells “know” this ahead of time and ensure that polarization sites were positioned next to (rather than overlapping) those sites?
In most fungal and animal cells, a polarity axis chosen in a cell type–specific manner is communicated to a conserved polarity mechanism centered on the Rho family GTPase Cdc42 (Johnson, 1999; Pruyne and Bretscher, 2000; Etienne-Manneville, 2004; Park and Bi, 2007). In S. cerevisiae, the polarity axis for budding is selected by a network of mating type–regulated bud-site selection proteins (Park and Bi, 2007). MATa or α cells bud axially (the new bud forms next to the previous division site) and MATa/α cells bud bipolarly (the new bud forms at either pole of the cell). Landmark proteins concentrated at the chosen sites recruit and activate Cdc24, the guanine nucleotide exchange factor (GEF) for Cdc42, which leads to localized production of GTP-Cdc42, which then directs cytoskeletal polarization and bud formation. However, whereas the landmark proteins localize to the site of cell division, Cdc24 and Cdc42 concentrate at an adjacent spot in preparation for budding. The basis for this avoidance of the previous division site was entirely unknown.
There are thought to be three dedicated GTPase-activating proteins (GAPs) for Cdc42 in yeast: Rga1, Rga2, and Bem3 (Zheng et al., 1994; Stevenson et al., 1995; Smith et al., 2002). All three are large proteins with C-terminal Rho GAP domains. Deletion of these GAPs either singly or in combination does not impair actin polarization but does impair Cdc42-directed assembly of the septin cytoskeleton into a ring at the presumptive bud site (Gladfelter et al., 2002; Smith et al., 2002; Caviston et al., 2003). Rga2 and Bem3 display a similar localization pattern to Cdc42 throughout the cell cycle. Although Rga1 also colocalizes with Cdc42 at the site of bud emergence, it subsequently spreads throughout the bud cortex and concentrates at the septin ring in the mother-bud neck until the end of the cell cycle (Caviston et al., 2003). Based on this unique localization pattern, we suspected that Rga1 might play a specialized role in addition to its shared roles with the other GAPs. Here, we show that Rga1 specifically prevents Cdc42 activation and thus budding within the old division site.
Results and discussion
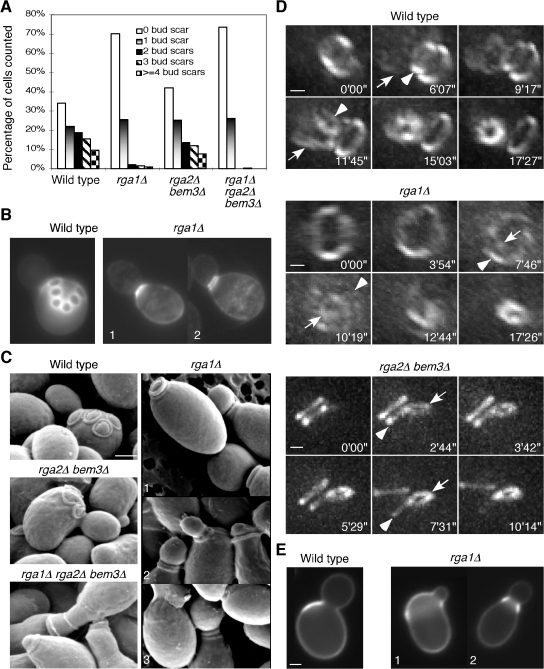
The most striking phenotype of rga1Δ single mutants is a paucity of bud scars (Fig. 1 A), which confirms previous findings (Stevenson et al., 1995; Chen et al., 1996). In contrast, rga2Δ bem3Δ cells displayed normal scar numbers (Fig. 1 A). The bud scar is a ridge of cell wall material formed by a septin-localized chitin synthase complex during bud emergence (DeMarini et al., 1997). The chitin ring stabilizes the bud neck during bud growth (Schmidt et al., 2003) and remains on the mother cell wall as a bud scar after cell division (Chant and Pringle, 1995). In principle, the dearth of bud scars in rga1Δ mutants could reflect fewer budding cycles, the ability to form buds without leaving scars, or the occurrence of multiple budding events at the same site. The first possibility predicts that rga1Δ populations would proliferate much more slowly than wild-type controls, but this was not observed (Chen et al., 1996). The second possibility seemed unlikely, given that all budded cells displayed bright Calcofluor-stained chitin rings on the mother side of the neck (Fig. 1 B). Thus, we tested whether new buds might form at the sites of old bud scars. Visualizing bud scars with scanning EM (SEM), we confirmed that wild-type haploid cells displayed nonoverlapping bud scars forming a zigzag line on the cell surface (Fig. 1 C). rga2Δ bem3Δ cells displayed a similar pattern, but rga1Δ cells often displayed more than one bud scar at the base of the bud neck (Fig. 1 C), which is suggestive of repeated budding at the same site. The rga1Δ rga2Δ bem3Δ cells also displayed stacked bud scars, though cells and scars were more irregular in shape (Fig. 1 C), as was expected given the perturbed septin rings in these mutants (Gladfelter et al., 2002; Smith et al., 2002; Caviston et al., 2003).
Figure 1.
Deletion of RGA1 causes polarization and budding within the previous division site. (A) Quantitation of bud scar distribution in an asynchronous population of cells from haploid strains YEF473A (wild type), YEF2324 (rga1Δ), YEF2392 (rga2Δ bem3Δ), and YEF2380 (rga1Δ rga2Δ bem3Δ). 200 cells were counted for each strain and unbudded daughter cells were excluded. The cells with only chitin rings at the base of the growing buds were counted as having “0 bud scar.” (B) Chitin staining of wild-type and rga1Δ cells indicated in A. Double chitin rings at the neck of an rga1Δ cell (2) were visualized occasionally when the distance between the rings was large enough to be resolved by light microscopy. (C) SEM observation of bud scars. The same strains described in A were used for SEM. (D) Using the positions of the septin rings as a read-out of the budding patterns in live cells. Cells of haploid strains YZT82 (CDC3-GFP, wild type), YZT55 (rga1Δ CDC3-GFP), and YZT111 (rga2Δ bem3Δ CDC3-GFP) were grown to exponential phase in YM-P medium and observed by 3D time-lapse microscopy at 30°C. Times are given in minutes and seconds after an arbitrary starting point. Arrowheads indicate an old septin ring at the mother side of the bud neck; arrows indicate the nascent septin ring at the new bud site. Views of the 3D images from particular angles are shown: an angled side view of the bud neck of the wild-type cell, an en-face view of the mother side of the bud neck of the rga1Δ cell, and a side view of the bud neck of the rga2Δ bem3Δ cell. Please note that a clear rotation of the mother cell versus the daughter cell occurred after cytokinesis and cell separation at a time between 9 min 17 s and 11 min 45 s for the wild-type cell and between 5 min 29 s and 7 min 31 s for the rga2Δ bem3Δcell. (E) The first bud of rga1Δ daughter cells forms within the birth scar. Birth scars of representative wild-type (YEF473A) and rga1Δ (YEF2324) cells. Cells 1 and 2 represent off-center and central budding within the birth scar, respectively. Bars, 1 μm.
To determine whether the stacked bud scars arose by repetitive budding from the same site, we performed 3D time-lapse microscopy on haploid cells expressing a functional GFP-tagged septin. Under optimal growth conditions, the new septin ring in a mother cell forms 5–8 min before the old septin ring at the previous division site disappears (Fig. 1 D; Iwase et al., 2006), providing an opportunity to visualize the new budding event with respect to the old division site. We found that wild-type (n = 7 of 8) and rga2Δ bem3Δ (n = 10 of 10) cells formed a new septin ring next to the disassembling old ring (Fig. 1 D). Strikingly, however, most rga1Δ cells (n = 22 of 30) and rga1Δ rga2Δ bem3Δ cells (n = 7 of 7) formed a new septin ring within the old ring (Fig. 1 D), which indicates that the stacked bud scars are indeed caused by repetitive budding from the same site. To determine where bud emergence occurs in rga1Δ daughter cells, we isolated newborn daughters by centrifugal elutriation, allowed them to form their first buds, and stained them with fluorescent Con A to visualize birth scars (Lew and Reed, 1993). Birth scars mark the sites on newborn cells that used to be the mother-bud neck and 96.5% of wild-type daughter cells (n = 231) form buds next to the birth scar (Fig. 1 E). Strikingly, 95.9% of rga1Δ daughter cells (n = 243) formed buds within (rather than next to) the birth scar (Fig. 1 E). This phenotype was confirmed by 3D time-lapse microscopy (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200705160/DC1). We also found that rga1Δ cells had expanded birth scars with a mean width of 3.8 ± 1.4 μm relative to wild-type birth scars of 1.6 ± 0.3 μm (mean ± standard deviation, n = 20), perhaps because of increased cell surface growth within the birth scar after cytokinesis and/or a mild defect in septin ring assembly. Together, these results demonstrate that Rga1 is necessary to prohibit budding at the previous division site in both mother and daughter cells.
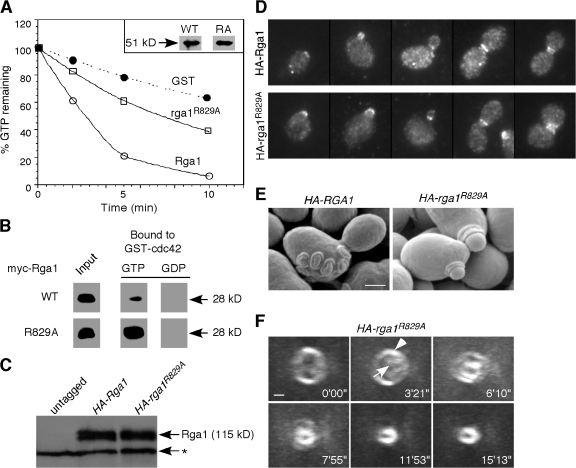
Previous studies have suggested that GAPs can potentially act not only as down-regulators but also as effectors for Cdc42. To determine whether Rga1 function requires GAP activity, we mutated the “arginine finger” motif conserved among Ras, Rho, and Cdc42 GAPs (Ahmadian et al., 1997; Rittinger et al., 1997). As expected, the Rga1R829A GAP domain displayed significantly reduced GAP activity toward Cdc42 in vitro (Fig. 2 A), though it retained the ability to bind GTP-Cdc42 (indeed, the mutant protein pulled down more GTP-Cdc42 than did the wild-type; Fig. 2 B). We then replaced the endogenous RGA1 with full-length epitope-tagged wild-type or R829A mutant genes. These proteins were expressed at comparable levels (Fig. 2 C) and localized similarly throughout the cell cycle (Fig. 2 D), but the arginine finger mutant was completely nonfunctional in terms of the budding pattern (Fig. 2, E and F; and Table I). Thus, the role of Rga1 in preventing polarization at the division site depends on its GAP activity.
Figure 2.
The role of Rga1 in polarity axis determination depends on its Cdc42-GAP activity. (A) GAP assays. Cdc42 prebound to γ-[32P]GTP was incubated with GST, the GST-Rga1 GAP domain, or the same domain containing the R829A mutation, and radioactivity remaining bound to Cdc42 is plotted against time of incubation. The inset shows that similar amounts of wild-type and mutant GAP domains were used in the assay. This GAP assay is representative of three experiments with consistent results. (B) Binding assays. Recombinant myc-tagged Rga1 or rga1R829A GAP domains were incubated with bead-bound recombinant GST-cdc42Q61L (GTP-bound) or GST-cdc42T17N (GDP-bound) to assess binding. (C) HA-Rga1 and HA-rga1R829A are expressed at similar levels. Protein samples were prepared from YZT194 (HA-RGA1) and YZT195 (HA-rga1R829A). The asterisk indicates a cross-reacting protein with the HA-antibody in yeast extracts. (D) HA-Rga1 and HA-rga1R829A display similar localization patterns in the cell cycle. Strains YZT194 and YZT195 were grown to exponential phase in YM-P medium at 23°C and examined by immunofluorescence using an anti-HA antibody. (E) SEM observation of bud scars of strains YZT194 and YZT195. (F) Visualization of new septin ring formation in live cells of strain YZT198 (HA-rga1R829A CDC3-GFP) by 3D time-lapse microscopy. Eight out of nine cells were observed to form a new septin ring within the old ring. The arrowhead indicates the old septin ring; the arrow indicates the new septin ring. Bars, 1 μm.
Table I. Bud scar distribution in cells containing different alleles of RGA1 or its GAP domain fused to BUD3, MLC2, or CYK3 .
| Strains | Percentage of cells with
|
||||
|---|---|---|---|---|---|
| Zero bud scars | One bud scar | Two bud scars | Three bud scars | Four bud scars | |
| HA-RGA1 | 40.0 | 20.0 | 12.5 | 11.0 | 16.5 |
| HA-rga1R829A | 72.5 | 22.5 | 4.0 | 1.0 | 0.0 |
| GFP-RGA1 | 38.0 | 24.5 | 12.5 | 16.5 | 8.5 |
| GFP-rga1700–1007aa rga1Δ∷HIS3 | 68.0 | 21.0 | 8.5 | 2.0 | 0.5 |
| rga1Δ∷HIS3 BUD3-GFP | 76.5 | 17.5 | 5.5 | 0.5 | 0.0 |
| rga1Δ∷HIS3 BUD3-rga1700–1007aa-GFP | 43.5 | 26.0 | 12.5 | 11.5 | 6.5 |
| rga1Δ∷HIS3 BUD3-rga1700–1007aa,R829A-GFP | 68.5 | 24.5 | 4.5 | 2.0 | 0.5 |
| rga1Δ∷HIS3 BUD3-rga2712–1009aa-GFP | 68.0 | 26.0 | 4.0 | 1.0 | 1.0 |
| rga1Δ∷HIS3 BUD3-bem3818–1128aa-GFP | 77.0 | 15.0 | 6.0 | 1.0 | 1.0 |
| rga1Δ∷HIS3 MLC2-GFP | 62.5 | 23.5 | 12.0 | 2.0 | 0.0 |
| rga1Δ∷HIS3 MLC2-rga1700–1007aa-GFP | 53.5 | 24.5 | 16.0 | 4.0 | 2.0 |
| rga1Δ∷HIS3 CYK3-GFP | 71.0 | 20.5 | 7.5 | 1.0 | 0.0 |
| rga1Δ∷HIS3 CYK3-rga1700–1007aa-GFP | 38.5 | 17.0 | 22.0 | 13.5 | 9.0 |
200 cells were counted for each strain, excluding unbudded daughter cells. The cells with only one chitin ring at the base of a growing bud were counted as having zero bud scars.
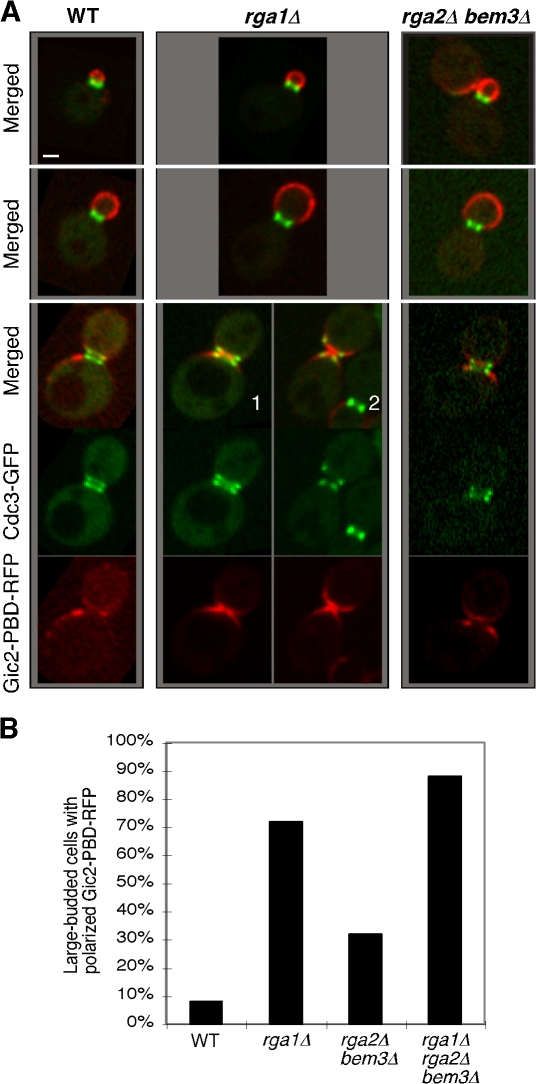
The budding-within-the-old-division-site phenotype and its dependency on the GAP activity of Rga1 raise the possibility that the level of GTP-Cdc42 may be elevated at the cell division site in rga1Δ cells. To examine this possibility, the localization of Gic2–p21 binding domain (PBD)–RFP, a reporter for GTP-Cdc42 (Fig. S1 B), was examined in wild-type and mutant strains (Fig. 3 and Videos 1–4, available at http://www.jcb.org/cgi/content/full/jcb.200705160/DC1). Gic2-PBD-RFP was localized to the presumptive bud site and the bud cortex of predivision cells in all strains. At the time when cells were completing cytokinesis and displayed split septin rings (detected with Cdc3-GFP), Gic2-PBD-RFP was localized adjacent to but outside the old division site in wild-type and most rga2Δ bem3Δ cells (Fig. 3, A and B; and Videos 1 and 4). In contrast, most rga1Δ and rga1Δ rga2Δ bem3Δ cells at this stage showed an increased concentration of Gic2-PBD-RFP within the split septin rings at the division site (Fig. 3, A and B; and Videos 2 and 3). This striking feature was confirmed by the localization of GFP-tagged full-length Gic2 and Ste20, effectors localized through their interaction with GTP-Cdc42 (Fig. S1 C). Thus, Rga1 is uniquely important for clearing GTP-Cdc42 from the bud neck at the end of cytokinesis.
Figure 3.
Deletion of RGA1 causes an elevated level of GTP-Cdc42 at the cell division site. (A) Cells of YZT292 (WT), YZT293 (rga1Δ), and YZT294 (rga2Δ bem3Δ) carrying integrated CDC3-GFP and GIC2-PBD-RFP were imaged by two-color light microscopy. Single representative GFP and RFP images from a stack of z sections for each cell were selected to show the localization patterns of Cdc3-GFP and Gic2-PBD-RFP with high resolution. Bar, 1 μm. (B) Quantitation of large-budded cells with neck-localized Gic2-PBD-RFP. Cells of YZT295 (rga1Δ rga2Δ bem3Δ) and other strains as in A were used and only large-budded cells with a clear septum (n = 50 for each strain) were scored.
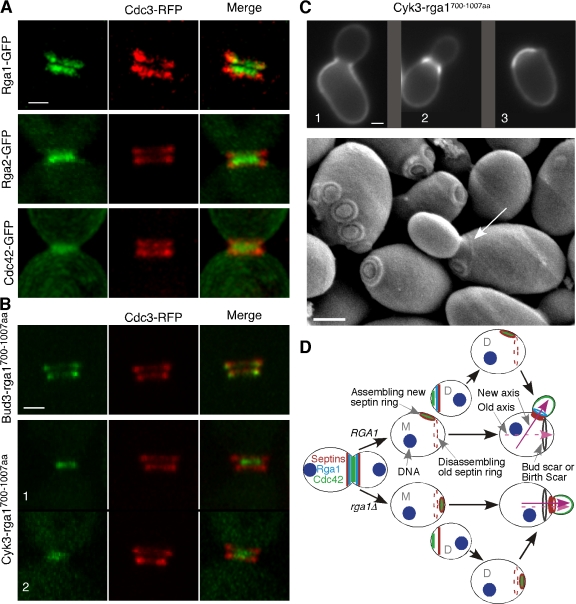
In addition to the C-terminal GAP domain, Rga1 has two N-terminal LIM domains. There is some evidence that these domains may restrict Rga1 GAP function, as their removal leads to synthesis of a truncated protein able to suppress morphogenesis defects caused by mutation of the Rho-GAP Bem2 (Chen et al., 1996). However, we found that deletion of the N-terminal half (or more) of Rga1 rendered the protein nonfunctional in terms of preventing polarization at the previous division site. The truncated protein was stably expressed (Fig. S1 E) but failed to concentrate at the cortical sites where full-length Rga1 was found (Fig. S1 F). Thus, the Rga1 non-GAP domains are important for both localization and function of Rga1. To determine whether proper localization of the Rga1 GAP domain to the septin rings at the end of the cell cycle was sufficient to prevent polarization at the previous division site, we fused the Rga1 GAP domain (residues 700–1007) to the bud-site selection protein Bud3, which is concentrated at the septin rings from G2 through cytokinesis and early G1 (Chant et al., 1995). Remarkably, this fusion was able to fully complement the same-site rebudding phenotype of rga1Δ mutants (Table I). Moreover, the GAP activity of the fusion protein was required because a Bud3-Rga1-GAPR829A mutant failed to restore function. Thus, the only essential role of the N-terminal 70% of Rga1 in preventing polarization at the previous division site is to ensure its localization to that site.
Why is it that only Rga1 (and not Rga2 or Bem3) can block polarization at the previous division site? We found that Rga1 and Rga2 displayed subtly different patterns of localization at the time of cytokinesis even though both concentrate at the mother-bud neck. In large-budded cells with split septin rings (i.e., cells undergoing cytokinesis), Rga1 was concentrated in two rings that lay within the two septin rings (visualized with a functional septin-DsRed.M1 fusion or septin-mCherry), whereas Rga2 was concentrated in a single patch that was either closer to the daughter side of the bud neck or sandwiched by two septin rings (Fig. 4 A). Total Cdc42 was also concentrated on the membranes predominantly appearing as a single patch that was sandwiched by two septin rings (Fig. 4 A). Thus, one possible basis for the functional specialization of Rga1 as compared with Rga2 is its unique localization within the septin rings during and after cytokinesis. To test this hypothesis, we first asked whether the Rga2 GAP domain (residues 712–1009) could block same-site rebudding when fused to Bud3. As with the Bud3-Rga1-GAP fusion described in the previous paragraph (Fig. 4 B), the Bud3-Rga2-GAP fusion protein localized to the septin rings before, during, and after cytokinesis (Fig. S1 G). However, this fusion failed to rescue the same-site rebudding defect (Table I). Similarly, a Bud3-Bem3-GAP fusion protein was unable to rescue the rga1Δ phenotype (Table I). These results suggest either that the Rga1 GAP domain has greater activity than the Rga2 and Bem3 GAP domains or that it plays an additional, unique role in excluding same-site rebudding.
Figure 4.
Targeting of the Rga1 GAP domain to the division site by heterologous proteins during and after cytokinesis is sufficient for the role of Rga1 in polarity-axis determination. (A) Fine patterns of Rga1, Rga2, and Cdc42 localization with respect to the septin rings during cytokinesis. Live cells of YZT211 (RGA1-GFP CDC3-DsRed), YZT166 (CDC3-mCherry) carrying plasmid pRS426-RGA2-GFP (the endogenous level of Rga2 was difficult to detect, thus, a high-copy plasmid carrying Rga2-GFP was used here; the pattern of Rga2 localization did not change with the high-copy plasmid, but the GFP signal was significantly improved), and YZT221 (CDC42-GFP CDC3-DsRed) were observed by 3D fluorescence microscopy at 23°C. All images in A and B are oriented such that the bud side is up. (B) Localization of Bud3-Rga1-GAP and Cyk3-Rga1-GAP fusions during cytokinesis. Cells of YZT240 (BUD3-rga1700–1007aa-GFP CDC3-mCherry) and YZT241 (CYK3-rga1700–1007aa-GFP CDC3-mCherry) were observed by 3D fluorescence microscopy as described for A. (C) Differential suppression of budding pattern defects by the Cyk3-Rga1-GAP fusion for the mother and daughter cells after cell division. (top) Birth scars of the strain JGY1645 (rga1Δ CYK3-rga1700–1007aa-GFP) were visualized. 1 and 2 represent off-center and central budding within the birth scar, respectively; 3 represents axial budding in a suppressed cell. (bottom) Birth scar (arrow) and bud scars of the strain JGY1645 were visualized by SEM. Strain JGY1645 was grown to exponential phase in YP medium containing 2% glycerol (YPG) and processed for SEM. The birth scar appeared to be better visualized in the YPG-poor medium than in the YPD-rich medium. Only 3.3% of the CYK3-RGA1-GAP mother cells (n = 273) budded within the bud scar when the strain was grown in YPG medium. (D) A model for the role of Rga1 in polarity-axis determination. New axis and Old axis refer to the axes of polarized cell growth. M, mother; D, daughter. Bars, 1 μm.
In a complementary approach to determine the importance of the precise septin-associated Rga1 localization, we fused the Rga1 GAP domain to the cytokinesis proteins Mlc2 and Cyk3. Mlc2 is the regulatory light chain of Myo1, the sole type II myosin in budding yeast, and it localizes to the bud neck from bud emergence to the end of the actomyosin contraction (Luo et al., 2004). Cyk3 accumulates in anaphase, localizes with the actomyosin contractile ring during cytokinesis, and then largely disappears (though two faint and fuzzy bands can sometimes be detected after actomyosin ring constriction; Korinek et al., 2000). Mlc2-Rga1-GAP and Cyk3-Rga1-GAP localized similarly to Mlc2 and Cyk3, respectively, thereby changing the Rga1 GAP localization to more closely resemble that of Rga2 and Bem3 during cytokinesis (Fig. 4 B, 1, during actomyosin ring contraction, and 2, presumed to be immediately after the actomyosin ring contraction; and not depicted). Mlc2-Rga1-GAP fusion failed to complement the rga1Δ phenotype (Table I). To our surprise, the Cyk3-Rga1-GAP fusion efficiently suppressed the same-site rebudding pattern defect of rga1Δ cells (Table I). SEM showed that only 1.5% of the cells (n = 197) carrying the Cyk3-Rga1-GAP fusion still budded within the old division site compared with 77% of the rga1Δ cells (n = 31). However, when we tested suppression of the daughter cell budding pattern, only 15.4% of the daughter cells (n = 273) budded axially, 52.7% still budded within the birth scar, and the rest were ambiguous (Fig. 4 C). The suppression of mother cell budding pattern and the failure to rescue the daughter cell budding pattern were clearly visualized in a single cell by SEM (Fig. 4 C, arrow indicates birth scar). The partial rescue of the daughter cell budding pattern was often associated with the rescue of the enlarged birth scar phenotype (the mean width of the birth scars for CYK3-rga1700–1007aa cells is 2.8 ± 1.1 μm, n = 20). We speculate that the Cyk3-Rga1-GAP protein was not effectively degraded at the end of cytokinesis in a subset of cells, allowing the remaining fusion protein to clear up the leftover GTP-Cdc42 within the birth scar, eliminating birth scar expansion, and blocking rebudding within the division site.
A major difference between mother and daughter cells is that daughter cells spend a longer time in G1 growing to the critical size before starting the next cell cycle (Johnston et al., 1977). Because Cyk3 is degraded at the end of mitosis, the simplest explanation for the ineffective rescue of the daughter cell budding pattern by the Cyk3-Rga1-GAP fusion protein is that, in daughter cells, the exclusion zone established by Cyk3-Rga1-GAP during cytokinesis has dissipated by the time that the cells initiate the next cell cycle and polarize. In contrast, mother cells begin the next cell cycle almost immediately after cytokinesis, when the exclusion zone is still in effect, so their budding pattern defect is effectively rescued by Cyk3-Rga1-GAP.
In aggregate, our findings indicate that the Rga1 GAP domain must be present at the division site to prevent subsequent polarization toward that site (Fig. 4 D). In daughter cells, which have a longer interval between division and subsequent polarization, it is also important for Rga1 GAP activity to persist after cytokinesis (Fig. 4 D).
We also investigated the role of Rga1 in diploid cells that bud in a bipolar pattern. Homozygous rga1Δ diploid cells also displayed the rebudding-within-the-old-division-site phenotype, but to a much lesser degree than rga1Δ haploids (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200705160/DC1). Deletion of BUD3, which causes bipolar budding in haploid cells (Chant et al., 1995), similarly decreased the percentage of rga1Δ haploid cells that budded within the old division sites (Fig. S2 B). This was expected because, unlike in axially budding cells where bud-site selection proteins always concentrate at the old division site, in bipolar budding, many cells concentrate these factors (and hence Cdc24) at the opposite pole. In such cells, we would not expect Rga1 to be needed. We also examined cells deleted for RSR1, which display a random budding pattern (Bender and Pringle, 1989). Here as well, the rga1Δ phenotype (while present) was quantitatively less penetrant (Fig. S2 B). These findings indicate that deletion of RGA1 causes same-site rebudding in all contexts but that the penetrance of the phenotype is most extreme in axially budding cells, where Rsr1 and associated bud-site selection proteins act to concentrate Cdc24 at the old division site.
Why is it important for yeast cells to avoid rebudding at the same site? In most wild yeast strains, daughter cells remain attached to their mothers for prolonged periods after cytokinesis (laboratory strains have been selected to detach rapidly to reduce clumping and make experimental manipulation easier). Indeed, in some circumstances cells need to remain robustly attached to penetrate solid substrates (e.g., during haploid invasive growth or diploid pseudohyphal growth; Pan et al., 2000; Breitkreutz and Tyers, 2002). Clearly, rebudding at the same site would be impossible if the previous daughter cell continued to occupy that space. Even in our laboratory strain, we observed that in some rga1Δ cells (n = 4 out of 30), new septin rings started to form within the old rings but were then aborted, and new rings then appeared at the opposite pole of the cell (Fig. S2 C). This behavior suggests that the previous division site, with its remnant bud scar, sometimes creates difficulties when attempting to rebud at that site, leading to aborted budding attempts. Therefore, the exclusion zone provided by Rga1 may have evolved to make budding more efficient by avoiding attempts to bud at difficult or occupied sites. Previous work indicated that axial bud-site selection proteins are deposited in a ring at the division site and subsequently recruit and activate Cdc24 (Park and Bi, 2007). Thus, Rga1 and Cdc24 establish concentric zones of negative and positive Cdc42 regulation that lead to the adjacent positioning of cellular structures.
For many families of small monomeric GTPases, there appear to be more GEFs and GAPs than there are G proteins (e.g., ∼53 GEFs and 68 GAPs for 17 Rho family GTPases in humans; Bernards, 2003; Bernards and Settleman, 2004). Our work demonstrates that one specific GAP is uniquely used to enforce an exclusion zone for cell polarization within a previous division site, which supports the hypothesis that GAPs play specialized roles. Moreover, the GAP must act at a specific location (the division site) and a specific time in the cell cycle (after cytokinesis). These findings are consistent with the hypothesis that the excess of regulators over G proteins evolved to exert exquisite spatiotemporal control over the activation of the G proteins, enabling each G protein to fulfill several cellular roles.
Materials and methods
Strains and growth conditions
Yeast strains used in this study are listed in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200705160/DC1). Standard culture media and genetic techniques were used as described previously (Guthrie and Fink, 1991). For some experiments, yeast was grown in YM-P (Lillie and Pringle, 1980), a rich, buffered liquid medium.
Construction of plasmids and yeast strains
Plasmids YEp181-HA-RGA1 and YEp181-HA-RGA1R829A were constructed as follows: an ∼5.8-kb HindIII fragment was subcloned from pALTER-1-HA-RGA1 (Caviston et al., 2003) into a HindIII site in YEp351 to generate YEp351-HA-RGA1. Then, an ∼3-kb XhoI–BglII fragment containing N-terminal HA-RGA1 was subcloned from YEp351-HA-RGA1 to replace the corresponding fragment in YEp181-RGA1 (Caviston et al., 2003) to generate YEp181-HA-RGA1. To generate YEp181-HA-RGA1R829A, an ∼1.5-kb AgeI–BglII fragment containing the R829A site from YEp181-RGA1R829A, which was constructed similarly to YEp181-RGA1 (K872A) (Caviston et al., 2003) except that the donor plasmid for the R829A mutation was pDLB1810, was used to replace the corresponding fragment in YEp181-HA-RGA1. Yeast strains YZT194 and YZT195 were constructed by integrating HindIII-digested YEp181-HA-RGA1 and YEp181-HA-RGA1R829A, respectively, into strain YZT88 (rga1Δ∷URA3-KanMX6) by homologous recombination. Plasmid YIp128-CDC3-DsRed.M1 (integrative, LEU2) was constructed by PCR amplifying DsRed.M1 as a 0.7-kb NotI fragment using plasmid pDsRed.M1 (provided by B. Glick, University of Chicago, Chicago, IL) as the template and the following pair of primers: DsRed-1F (5′-ATAAGAATGCGGCCGC ATGGACAACACCGAGGAC-3′; the underlined sequence represents the NotI site and the bold sequence represent the 5′ end coding region of DsRed.M1) and DsRed-1R (5′-TAGTTTAGCGGCCGCAggatccCTGGGAGCCGGAGTGGCG-3′; the underlined sequence represents the NotI site, the lower case letters represent the BamHI site, and the bold sequence represents the 3′ end coding region of DsRed.M1, excluding the stop codon). The NotI fragment carrying DsRed.M1 was used to replace the NotI-GFP cassette in YIp128-CDC3-GFP in an appropriate orientation, resulting in the desired plasmid. Plasmid YIp128-CDC3-mCherry (integrative, LEU2) was constructed similarly to YIp128-CDC3-DsRed.M1 except that the mCherry-containing plasmid pKT355 (supplied by K. Thorn, University of California, San Francisco, San Francisco, CA; Sheff and Thorn, 2004; Iwase et al., 2006) was used as the template and the following primers were used: mCherry-NotI-F (5′-ATAAGAATGCGGCCGC ATGGTGAGCAAGGGCGAGGAG-3′; the underlined sequence represents the NotI site and the bold sequence represents the 5′ end coding region of mCherry) and mCherry-NotI-R (5′-ATAAGAATGCGGCCGCAggatccCTTGTACAGCTCGTCCATGCC-3′; the underlined sequence represents NotI site, the lower case letters represent the BamHI site, and the bold sequence represents the 3′ end coding region of mCherry, excluding the stop codon).
To generate plasmid pRS426-RGA2-GFP-KanMX6 (2 μm URA3) for the localization experiment, a PCR product was amplified from pFA6a- GFP(F64L/S65T)-KanMX6 (Longtine et al., 1998) using RGA2-2 forward and RGA2-R1 reverse primers. The PCR product encoding the GFP- KanMX6 cassette with an RGA2 C-terminal coding sequence excluding the stop codon and the 3′ untranslated region was transformed into wild-type cells harboring plasmid pDLB1981 (pRS426-RGA2; Gladfelter et al., 2002) and KanR colonies were selected to yield plasmid pRS426-RGA2- GFP-KanMX6.
To generate plasmid pRS306-BUD3-C-GFP for the Bud3–C-terminal fusion experiment, a PCR-amplified BamHI–EcoRI fragment encoding aa 1477–1636 of Bud3 without a stop codon was inserted into the pRS306-TCYC1 vector (integrative, URA3) to generate pRS306-BUD3-C-TCYC1. Then, a unique MluI site within TCYC1 of pUG35 (CEN LEU2 MET25p-yEGFP; supplied by J.H. Hedgemann, Heinrich-Heine-Universität, Düsseldorf, Germany) was destroyed by MluI digestion followed by filling-in with the Klenow fragment of DNA polymerase I to generate pUG35-ΔMluI. The EcoRI–KpnI fragment of yEGFP3-TCYC1 from pUG35-ΔMluI was inserted into EcoRI- and KpnI-digested pRS306-BUD3-C-TCYC1 to generate pRS306-BUD3-C-GFP.
The HindIII–SalI DNA fragment containing RGA1-C1 (encoding aa 700–1007) was amplified by PCR from Yep181-HA-RGA1, digested with HindIII and SalI, and ligated into a HindIII- and SalI-digested pRS306-BUD3-C-GFP vector to generate pRS306-BUD3-RGA1-C1-GFP. This plasmid, along with pRS306-BUD3-C-GFP, was linearized with MluI for integration at the BUD3 locus of the strain YEF2324 (rga1Δ) cells to generate strains JGY1622 and JGY1621. Similarly, the HindIII–SalI DNA fragment containing RGA1-C1 (aa 700–1007, R829A) was amplified by PCR from YEp181-HA-RGA1R829A, digested with HindIII and SalI, and ligated into a HindIII- and SalI-digested pRS306-BUD3-C-GFP vector to generate pRS306-BUD3-RGA1-C1-GFP (aa 700–1007, R829A). The plasmid was linearized with MluI for integration at the BUD3 locus of the strain YEF2324 (rga1Δ) cells to generate strain JGY1639.
To generate Bud3-Rga2-GAP or Bud3-Bem3-GAP fusion, the HindIII–SalI DNA fragments containing RGA2-C1 (encoding aa 712–1009) or BEM3-C1 (encoding aa 818–1128) were amplified by PCR, digested with HindIII and SalI, and ligated into a HindIII- and SalI-digested pRS306-BUD3-C-GFP vector to generate pRS306-BUD3-RGA2712–1009aa-GFP and pRS306-BUD3-BEM3818–1128aa-GFP. The later two plasmids were linearized with MluI for integration at the BUD3 locus of strain YEF2324 (rga1Δ) cells to generate strains JGY1641 and JGY1642.
To generate plasmid pRS306-CYK3-C-GFP (integrative, URA3) for the Cyk3–C-terminal fusion experiment, the EcoRI-KpnI fragment of yEGFP3-TCYC1 from pUG35-ΔMluI was ligated into a EcoRI- and KpnI- digested pRS306 vector. Then, a PCR-amplified XbaI–BamHI fragment encoding aa 564–885 of Cyk3 without a stop codon was inserted into XbaI- and BamHI-digested pRS306-yEGFP3-TCYC1 to generate pRS306-CYK3-C-GFP. Plasmid pRS306-RGA1-C1-yEGFP3 was constructed by inserting the HindIII–KpnI fragment of RGA1-C1-yEGFP3-TCYC1 from pRS306-BUD3-RGA1-C1-GFP into pRS306. The SacI–BamHI fragment of CYK3-C from pRS306-CYK3-C-GFP was inserted into SacI- and BamHI-digested pRS306-RGA1-C1-yEGFP3 to generate pRS306-CYK3-RGA1-C1-GFP. Plasmids pRS306-CYK3-C-GFP and pRS306-CYK3-RGA1-C1-GFP were linearized with BglII and ClaI, respectively, for integration at the CYK3 locus of strain YEF2324 (rga1Δ) cells to generate strains JGY1644 and JGY1645. For this purpose, pRS306-CYK3-RGA1-C1-GFP had to be prepared in an Escherichia coli host strain with a dam mutation in order for the normally methylated unique ClaI site to be digested.
To generate strains carrying MLC2-GFP or MLC2-rga1-GAP-GFP fusion, a PCR-amplified fragment encoding full-length Mlc2 (aa 1–163) without a stop codon was digested with SacI and SpeI and ligated into SacI- and SpeI-digested pRS306-yEGFP3-TCYC1 and pRS306-RGA1-C1-yEGFP3 to generate pRS306-MLC2-C-GFP and pRS306-MLC2-RGA1-C1-GFP. The latter two plasmids were linearized with SacII for integration at the MLC2 locus on the chromosome in an rga1Δ strain (YEF2324) to generate strains YZT283 and YZT284.
To generate the RGA1-C1-GFP (encoding aa 700–1007) fusion construct, a 2.4-kb KpnI–BamHI fragment containing a 1.17-kb RGA1 promoter, an ATG start codon, and the GFP sequence from pRS315-GFP-RGA1 (Caviston et al., 2003) was ligated into a YIplac211 vector (integrative, URA3) to generate YIp211-PRGA1-GFP. This new plasmid was digested with BamHI and HindIII and the RGA1-C1 fragment was inserted into this plasmid to generate YIp211-RGA1-C1. This plasmid was linearized with a unique XhoI site within the RGA1 promoter region for integration at the endogenous RGA1 locus of the strain YEF2324 (rga1Δ) cells to generate strain YZT232.
The plasmid YIp211-GIC2-PBD-RFP (integrative, URA3) was constructed as follows: a pair of hybrid primers (GIC2-F5-208-RFP, TCGTGAAAGCAAACTCATTTCAAGATCGCACGAAAATAAGGGTGACGGTGCTGGTTTA, and GIC2-R3-RFP, ACGTTACTGAGATCGAACGCGCGACTGATAGTCTTGATGTTCGATGAATTCGAGCTCG) were used to amplify tdTomato-SpHIS5 from the template plasmid (pKT356, also called pFA6a-link-tdTomato-SpHIS5; supplied by K. Thorn) and the PCR fragment was transformed into yeast strain YEF3967 (gic2Δ∷KanMX; Invitrogen) carrying plasmid pCC967 (2 mm, URA3, GIC2 under its own promoter control; Bi et al., 2000). Plasmids were recovered from His+ Ura+ yeast transformants and confirmed to contain appropriate GIC2-PBD-RFP fusion by PCR checking using a pair of checking primers (a 20-bp forward primer that is 420 bp downstream of the GIC2 start codon TCTCCACACCATTTGATTTT and a 20-bp reverse primer that is 104 bp upstream of the GIC2 stop codon GATTGTGGAGAAGGCGTAGC). In addition, the junction between the GIC2 sequence and tdTomato in the fusion constructs has been sequence confirmed. From this procedure, two fusion constructs were obtained, pCC967-tdTomato and pCC967-1.5tdTomato, both of which contain an in-frame fusion between the first 208 codons of GIC2 encoding the PBD or the Cdc42/Rac interactive binding domain and the tdTomato sequence, except that latter plasmid contained one and a half copies of tdTomato (essentially three copies of RFP in tandem), which was formed presumably by recombination in yeast between GIC2 on plasmid pCC967 and two copies of a PCR fragment carrying tdTomato-SpHIS5. A SacI fragment carrying GIC2-PBD-RFP was isolated from both plasmids (∼3.9 kb for pCC967-tdTomato and ∼4.8 kb for pCC967-1.5tdTomato) and cloned into the integrative plasmid YIplac211 (integrative, URA3) at the SacI site, resulting in two plasmids, YIp211-GIC2-PBD-tdTomato and YIp211-GIC2-PBD-1.5tdTomato, respectively. Both plasmids were linearized by ApaI digestion and integrated at the ura3 locus of a yeast strain for localization comparison. Both integrated fusion constructs yielded the same pattern of localization, except that the Gic2-PBD-1.5tdTomato signal was brighter. Thus, YIp211-GIC2-PBD-1.5tdTomato was used throughout this study and, for simplicity, this plasmid was renamed YIp211-GIC2-PBD-RFP.
SEM
Cells were prepared by fixation, dehydration, critical point drying, and sputter coating with gold-palladium as described previously (Chant and Pringle, 1995). Cells were observed and photographed digitally using an FEI XL-20 SEM (Philips).
3D time-lapse microscopy
Cells and slides were prepared as described previously (Iwase et al., 2006). Cells carrying CDC3-GFP were imaged at intervals of 2–5 min using a microscope system (DeltaVision Spectris; Applied Precision) and a charge-coupled device camera (Cool-Snap HQ; Roper Scientific) to follow the disassembly of the old septin ring and the formation of the new septin ring at and adjacent to the bud neck. For each time point, 30 images were acquired at 0.3-μm increments, deconvolved, and reconstructed into a 3D image. Imaging was performed at 30°C.
For the two-color localization experiments presented in Figs. 3 A and 4 (A and B), images of live cells were acquired digitally by the MetaMorph-controlled (MDS Analytical Technologies) TE2000 microscope (Nikon) equipped with a Plan Apo 100× 1.45 NA total internal reflection fluorescence oil immersion objective lens (Nikon), a Yokogawa spinning disk confocal scanner (PerkinElmer), and a deep-cooled ORCA II-ER charge-coupled device camera (Hamamatsu). The 488- and 568-nm laser lines of an argon/krypton laser (Melles Griot) were used for excitation of GFP and RFP in combination with a triple-band pass dichroic mirror. For each cell, 11 images of GFP and RFP at 0.3-μm increments for Fig. 3 A and 20 images of GFP and RFP at 0.2-μm increments for Fig. 4 (A and B) were acquired at 23°C.
Indirect immunofluorescence and bud scar staining
For localization of HA-Rga1 and HA-Rga1R829A, yeast cells grown exponentially in YM-P media at 24°C were fixed by formaldehyde and processed for immunofluorescence microscopy as described preciously (Pringle et al., 1991). A mouse monoclonal anti-HA primary antibody (HA.11; Covance) and a secondary Cy3-conjugated donkey anti–mouse IgG antibody (Jackson ImmunoResearch Laboratories) were used. Differential interference contrast and fluorescence microscopy were performed using a microscope (E800; Nikon) with a 60× Plan Apo objective. The images were acquired using Image-Pro Plus software (Media Cybernetics, Inc.).
Bud scars were visualized by fluorescence microscopy with the E800 microscope after staining with Calcofluor (Sigma-Aldrich). Cells were fixed by the addition of formaldehyde to 3.7% and incubation for ∼2 h with occasional agitation. Cells were then stained with 0.1% Calcofluor as described previously (Pringle, 1991).
Centrifugal elutriation and birth scar staining
Enrichment of small daughter cells from exponentially growing cultures was achieved by centrifugal elutriation as described previously (Lew and Reed, 1993). After elutriation, cells were grown in rich medium YEPD at 30°C for 100–160 min (100 min after elutriation for the wild-type and rga1Δ cells and 160 min for the CYK3-rga1-GAP cells). Samples were fixed with 3.6% formaldehyde for 2 h at room temperature, washed with 0.1 M KPO4, pH 7.5, and resuspended in immunofluorescence solution B (0.1 M KPO4, pH 7.5, and 1.2 M sorbitol). Birth scars were stained with 12.5 μg/ml Alexa 594–ConA (Invitrogen) in immunofluorescence solution B for 20 min. Cells were examined using an AxioImager.A1 (Carl Zeiss, Inc.) with a 100× oil immersion objective. Images were captured using an ORCA cooled charge-coupled device camera and interfaced with MetaMorph software. Images were processed for presentation using Photoshop (Adobe). The means of the width of the birth scars in the post-elutriation cells of the rga1Δ, CYK3-rga1700–1007aa, and wild-type strains were determined by tracing the birth scar in cross section and the length of the traced line was determined by MetaMorph.
Protein assays
Production of recombinant proteins and GAP assays.
Production of GST-tagged proteins from E. coli and measurement of the GAP activity were all performed as described previously (Gladfelter et al., 2002). To determine the amount of recombinant GST-GAP domain to add to the assay, we first got an approximate estimate using the Bradford assay and then ran 1.5, 2, or 3 μl of wild-type GAP domain to compare to a single amount of mutant GAP domain on a Western blot to fine-tune the amount. Fig. 2 A shows the relevant lanes from that Western blot spliced next to each other using Photoshop. Similar Western-based quantitation was used to ensure that equal amounts of fusion proteins were used for the pull-down assays shown in Fig. 2 B.
Immunoblotting.
For immunoblotting of HA-Rga1 and HA-Rga1R829A, proteins were separated by 7% SDS-PAGE and transferred to nitrocellulose for blotting. A mouse monoclonal anti-HA primary antibody HA.11 and a secondary horseradish peroxidase–conjugated goat anti–mouse IgG (Jackson ImmunoResearch Laboratories) were used, and the HA-tagged proteins were detected using an ECL system (Millipore).
Online supplemental material
Fig. S1 shows budding within the birth scar, failure of Gic2-PBD-RFP localization in the Cdc42 GEF mutant cdc24-4, an increased level of Cdc42-GTP in rga1Δ cells as reported by Gic2-GFP and Ste20-GFP, budding within the bud scar in a hyperactive cdc42 mutant, expression levels of full-length Rga1 and Rga1-GAP domain, and localization of Rga1-GAP domain and Bud3-Rga2-GAP fusion. Fig. S2 shows the rga1Δ phenotype in diploid cells, the effects of deletion of bud-site selection genes on the rga1Δ phenotype in haploid cells, and an aborted attempt to bud within the old division site. Video 1 shows Cdc42 activation during cytokinesis and cell separation in a wild-type cell. Video 2 shows Cdc42 activation during cytokinesis and cell separation in an rga1Δ cell. Video 3 shows Cdc42 activation during cytokinesis and cell separation in an rga1Δ cell. Video 4 shows Cdc42 activation during cytokinesis and cell separation in an rga2Δ bem3Δ cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200705160/DC1.
Supplemental Material
Acknowledgments
We thank Steve Kane for making the Gic2-PBD-RFP construct, Drs. Ben Glick, Johannes Hedgemann, Hans-Ulrich Mösch, and Kurt Thorn for plasmids, Mr. Ray Meade from the Biomedical Imaging Core Laboratory at the University of Pennsylvania for his excellent technical assistance in SEM, Dr. Phong Tran for helping us with spinning-disc microscopy, Drs. Anna Kashina and Phong Tran for their comments on the manuscript, and the members of the Bi and Lew laboratories for discussions.
This work is supported by National Institutes of Health grants GM59216 to E. Bi and GM62300 to D.J. Lew.
Abbreviations used in this paper: GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; PBD, p21 binding domain; SEM, scanning EM.
References
- Ahmadian, M.R., P. Stege, K. Scheffzek, and A. Wittinghofer. 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat. Struct. Biol. 4:686–689. [DOI] [PubMed] [Google Scholar]
- Barton, A.A. 1950. Some aspects of cell division in Saccharomyces cerevisiae. J. Gen. Microbiol. 4:84–86. [DOI] [PubMed] [Google Scholar]
- Bender, A., and J.R. Pringle. 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras–related gene RSR1. Proc. Natl. Acad. Sci. USA. 86:9976–9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards, A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 1603:47–82. [DOI] [PubMed] [Google Scholar]
- Bernards, A., and J. Settleman. 2004. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 14:377–385. [DOI] [PubMed] [Google Scholar]
- Bi, E., J.B. Chiavetta, H. Chen, G.-C. Chen, C.S.M. Chan, and J.R. Pringle. 2000. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol. Biol. Cell. 11:773–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz, A., and M. Tyers. 2002. MAPK signaling specificity: it takes two to tango. Trends Cell Biol. 12:254–257. [DOI] [PubMed] [Google Scholar]
- Caviston, J.P., M. Longtine, J.R. Pringle, and E. Bi. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J., and J.R. Pringle. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129:751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J., M. Mischke, E. Mitchell, I. Herskowitz, and J.R. Pringle. 1995. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129:767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.-C., L. Zheng, and C.S.M. Chan. 1996. The LIM domain-containing Dbm1 GTPase-activating protein is required for the normal cellular morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:1376–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini, D.J., A.E.M. Adams, H. Fares, C. De Virgilio, G. Valle, J.S. Chuang, and J.R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville, S. 2004. Cdc42–the centre of polarity. J. Cell Sci. 117:1291–1300. [DOI] [PubMed] [Google Scholar]
- Gladfelter, A.S., I. Bose, T.R. Zyla, E.S.G. Bardes, and D.J. Lew. 2002. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J. Cell Biol. 156:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, San Diego. 933 pp.
- Iwase, M., J. Luo, S. Nagaraj, M. Longtine, H.B. Kim, B.K. Haarer, C. Caruso, Z. Tong, J.R. Pringle, and E. Bi. 2006. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 17:1110–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, D.I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston, G.C., J.R. Pringle, and L.H. Hartwell. 1977. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 105:79–98. [DOI] [PubMed] [Google Scholar]
- Korinek, W.S., E. Bi, J.A. Epp, L. Wang, J. Ho, and J. Chant. 2000. Cyk3, a novel SH3-domain protein, affects cytokinesis in yeast. Curr. Biol. 10:947–950. [DOI] [PubMed] [Google Scholar]
- Lew, D.J., and S.I. Reed. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie, S.H., and J.R. Pringle. 1980. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol. 143:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., A. McKenzie III, D.J. DeMarini, N.G. Shah, A. Wach, A. Brachat, P. Philippsen, and J.R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Luo, J., E.A. Vallen, C. Dravis, S.E. Tcheperegine, B. Drees, and E. Bi. 2004. Identification and functional analysis of the essential and regulatory light chains of the only type II myosin Myo1p in Saccharomyces cerevisiae. J. Cell Biol. 165:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, R.K., and J.R. Johnson. 1959. Life span of individual yeast cells. Nature. 183:1751–1752. [DOI] [PubMed] [Google Scholar]
- Pan, X., T. Harashima, and J. Heitman. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3:567–572. [DOI] [PubMed] [Google Scholar]
- Park, H.O., and E. Bi. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle, J.R. 1991. Staining of bud scars and other cell wall chitin with Calcofluor. Methods Enzymol. 194:732–735. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., A.E.M. Adams, D.G. Drubin, and B.K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565–602. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 113:365–375. [DOI] [PubMed] [Google Scholar]
- Rittinger, K., P.A. Walker, J.F. Eccleston, K. Nurmahomed, D. Owen, E. Laue, S.J. Gamblin, and S.J. Smerdon. 1997. Crystal structure of a small G protein in complex with the GTPase-activating protein rhoGAP. Nature. 388:693–697. [DOI] [PubMed] [Google Scholar]
- Schmidt, M., A. Varma, T. Drgon, B. Bowers, and E. Cabib. 2003. Septins, under Cla4p regulation, and the chitin ring are required for neck integrity in budding yeast. Mol. Biol. Cell. 14:2128–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff, M.A., and K.S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 21:661–670. [DOI] [PubMed] [Google Scholar]
- Smith, G.R., S.A. Givan, P. Cullen, and G.F. Sprague Jr. 2002. GTPase-activating proteins for Cdc42. Eukaryot. Cell. 1:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, B.J., B. Ferguson, C. De Virgilio, E. Bi, J.R. Pringle, G. Ammerer, and G.F. Sprague Jr. 1995. Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 9:2949–2963. [DOI] [PubMed] [Google Scholar]
- Zheng, Y., R. Cerione, and A. Bender. 1994. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J. Biol. Chem. 269:2369–2372. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.