Abstract
One-nanosecond molecular dynamics trajectories of three haloalkane dehalogenases (DhlA, LinB, and DhaA) are compared. The main domain was rigid in all three dehalogenases, whereas the substrate specificity-modulating cap domains showed considerably higher mobility. The functionally relevant motions were spread over the entire cap domain in DhlA, whereas they were more localized in LinB and DhaA. The highest amplitude of essential motions of DhlA was noted in the α4′-helix-loop-α4-helix region, formerly proposed to participate in the large conformation change needed for product release. The highest amplitude of essential motions of LinB and DhaA was observed in the random coil before helix 4, linking two domains of these proteins. This flexibility is the consequence of the modular composition of haloalkane dehalogenases. Two members of the catalytic triad, that is, the nucleophile and the base, showed a very high level of rigidity in all three dehalogenases. This rigidity is essential for their function. One of the halide-stabilizing residues, important for the catalysis, shows significantly higher flexibility in DhlA compared with LinB and DhaA. Enhanced flexibility may be required for destabilization of the electrostatic interactions during the release of the halide ion from the deeply buried active site of DhlA. The exchange of water molecules between the enzyme active site and bulk solvent was very different among the three dehalogenases. The differences could be related to the flexibility of the cap domains and to the number of entrance tunnels.
Keywords: a/b-hydrolase, catalytic triad, solvent mobility, specificity, structure-function, essential molecular dynamics
Haloalkane dehalogenases are bacterial enzymes cleaving the carbon-halogen bond of halogenated aliphatic compounds by hydrolysis (Janssen et al. 1994). These enzymes represent an attractive target for protein engineering studies attempting to improve their catalytic efficiency and broaden their substrate specificity toward important environmental pollutants (Copley 1998).
Initial classification of haloalkane dehalogenases into two classes was based on biochemical characteristics (Slater et al. 1995). Quantitative classification was based on statistical analysis of the activity data (Damborsky et al. 1997b; Nagata et al. 1997) and revealed the presence of three unique specificity classes within this protein family. Three different genes of haloalkane dehalogenases, corresponding to three proposed specificity classes, were subsequently detected in various bacteria isolated from contaminated localities worldwide (Poelarends et al. 2000). The "static" structural determinants of the substrate specificity were implied from the crystallographic analysis of three dehalogenases belonging to different specificity classes, that is, Xanthobacter autotrophicus GJ10 (DhlA; Verschueren et al. 1993b), Rhodococcus sp. (DhaA; Newman et al. 1999), and Sphingomonas paucimobilis UT26 (LinB; Marek et al. 2000). The substrate specificity appears to be modulated not only by the size and shape of the active site but also by the position and shape of tunnels connecting buried active sites with a bulk solvent (Damborsky and Koca 1999).
Haloalkane dehalogenases are composed of two domains: a main domain and a cap domain. The main domain is common to all haloalkane dehalogenases, as well as to many hydrolytic enzymes classified as α/β-hydrolases (Ollis et al. 1992; Nardini and Dijsktra 1999). The core of the main domain consists of an eight-stranded β-pleated sheet with seven parallel and one antiparallel strand surrounded by α-helices. The cap domain lies on the top of the main domain and consists of five helices connected by loops. Four of these helices, α4 through α7, resemble the structure of uteroglobin (Russell and Sternberg 1997). Two domains contribute to the formation of an internal cavity. This cavity is predominantly composed of hydrophobic residues. The only charged residues in the cavity are two of three catalytic residues: a nucleophile (aspartic acid) and a base (histidine). The third member of the catalytic triad is a catalytic acid (aspartic or glutamic acid). The characteristic structural motif of haloalkane dehalogenases is a halide-binding site. The halide ion released during the dehalogenation reaction is mainly stabilized by the hydrogen bonds provided by two tryptophan residues (Verschueren et al. 1993b) or by tryptophan and asparagine (Newman et al. 1999; Marek et al. 2000).
In the current study, an attempt is made to uncover "dynamic" structural determinants of substrate specificity of the haloalkane dehalogenases. The study extends previous molecular dynamics simulations conducted with the haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. Arnold and Ornstein (Arnold and Ornstein 1997) performed 300-ps molecular dynamics simulations with the free enzyme, the enzyme in the complex with substrate 1,2-dichloroethane, and the enzyme with the substrate trapped in covalent intermediate. Lightstone and coworkers (Lightstone et al. 1998) compared 540-ps molecular dynamics simulations of ground and transition states for the SN2 displacement of chloride ion from 1,2-dichloroethane in the enzyme active site.
One-nanosecond trajectories of the three haloalkane dehalogenases are compared here on the level of tertiary structures, secondary structure elements, amino acid residues, and active-site water molecules. The observed differences are discussed in light of the structure, function, and evolution of these enzymes.
Results
Characteristics of molecular dynamics trajectories
The initial potential energy of solvated protein structures (−1.2 • 105 to −1.6 • 105 kcal • mol−1) indicates that the systems were sufficiently minimized before simulations. The total energy reached a steady state after ∼50 ps and the mean energy values are reported in Table 1. The root mean square deviations (RMSDs), with respect to the crystal structures for all backbone atoms, reached the plateau of ∼1.0 Å in the first 50 ps of the simulations (Fig. 1 ▶). All atom RMSDs for DhlA reached the plateau of ∼1.5 Å, which is comparable to the value reported by Arnold and Ornstein (1997). The mean radius of gyration (Rg) calculated from the DhlA trajectory (18.18 ± 0.07 Å) is somewhat larger than that derived for the crystal structure (17.94 Å) but is smaller than the mean Rg value (18.40 Å) published for DhlA by Arnold and Ornstein (Arnold and Ornstein 1997). The mean Rg values for LinB and DhaA are also slightly larger than the mean Rg values calculated for the corresponding crystal structures (Table 1). Both mean Rg values and the solvent accessible surfaces (SASs) are stable during the simulation (Fig. 1 ▶). The mean SASs along the trajectory are not significantly different from the SASs of the crystal structures (Table 1). Comparison of selected parameters derived from the molecular dynamics trajectories with those calculated for crystal structures shows that the simulations are convergent and that the crystal structure geometries are well preserved during the simulations.
Table 1.
Summary of molecular dynamics trajectory characteristics
| Protein | E | Rg (MD) | Rg (X-ray) | SAS (MD) | SAS (X-ray) | RMSD |
| 105 kcal • mol−1 | Å | Å | 104 Å2 | 104 Å2 | Å | |
| DhlA | −0.98 ± 0.00 | 18.18 ± 0.07 | 17.94 | 1.31 ± 0.02 | 1.26 | 0.9 ± 0.1 |
| LinB | −1.02 ± 0.03 | 17.86 ± 0.05 | 17.42 | 1.21 ± 0.02 | 1.17 | 0.8 ± 0.1 |
| DhaA | −1.22 ± 0.00 | 17.96 ± 0.03 | 17.78 | 1.35 ± 0.01 | 1.20 | 1.0 ± 0.1 |
| SAS (phobic) | SAS (phylic) | α-helix | β-sheet | 310-helix | β-bridge | |
| 103 Å2 | 103 Å2 | |||||
| E, mean total energy; Rg (MD), mean radius of gyration; Rg (X-ray), radius of gyration of the X-ray structure; SAS (MD), mean solvent accessible surface; SAS (X-ray), solvent accessible surface of the X-ray structure; RMSD, root mean square deviation of backbone atoms with respect to the X-ray structure; SAS (phobic), solvent phobic part of the solvent accessible surface; SAS (phylic), solvent phylic part of the solvent accessible surface; α-helix, number of residues in α-helix; β-sheet, number of residues in β-sheet; 310-helix, number of residues in 310-helix; β-bridge, number of residues in β-bridge; all means were sampled and averaged along trajectory. | ||||||
| DhlA | 3.6 ± 0.7 | 9.6 ± 1.3 | 103.4 ± 3.9 | 44.9 ± 1.9 | 20.9 ± 3.6 | 0.0 ± 0.2 |
| LinB | 3.9 ± 0.9 | 8.4 ± 0.1 | 102.3 ± 4.6 | 52.7 ± 2.6 | 19.7 ± 4.0 | 3.2 ± 0.8 |
| DhaA | 4.5 ± 0.8 | 8.1 ± 0.9 | 105.9 ± 5.2 | 52.8 ± 1.9 | 18.7 ± 4.5 | 3.0 ± 0.4 |
Fig. 1.
Changes in the trajectory characteristics during the simulation. The root mean square deviation (in Å), radius of gyration (in Å), and solvent-accessible surface (in 104 Å2) are plotted against time (in picoseconds). The thermalization phase of simulations is not included in the graphs.
Dynamics of the overall protein structures
The key motions of the protein structures during the simulation were studied by the essential dynamics method (Amadei et al. 1993). This technique separates concerted, functionally relevant motions of a protein from Gaussian, physically constrained fluctuations using the analysis of covariance. The first two eigenvectors from this analysis covered most of the concerted motions in all three trajectories and will be discussed in detail.
The first concerted motion of DhlA starts in the loop before helix α4′, continues through helix α4 and helix α5, and ends up at the helix α6 (Fig. 2 ▶). The maximum amplitudes are observed around the residue Asp170, in the loop between helix α4′ and helix α4 and around the residue Arg193 in helix α5. The motion of residue Asp170 is transmitted to the loop between sheet β7 and helix α10 because of the existence of the salt bridge between Asp170 and Lys261. The salt bridge is formed between Asp170-Oδ1. . .Nζ-Lys261 and remains for 87.4% of the trajectory with a mean distance of 2.8 Å. The salt bridge breaks at 961 ps. The second eigenvector relates to more localized motions of the region between the residues Tyr31 and Leu34 in the loop between strand β1 and strand β2 and between the residues Leu151 and Gln158 in the loop before helix α4′.
Fig. 2.

Ensemble of 10 structures (in stereo pairs) of the first and the second essential motion obtained from the essential dynamics analysis. Only alpha-trace is shown. The most flexible residues and secondary elements are highlighted.
The first essential motion of LinB is localized on the random coil before helix α4, helix α4 and helix α5′, and the loop after helix α5′. The maximum amplitude occurs at the end of helix α4 and is probably caused by the stretching of helix α5′ (Fig. 2 ▶). The second essential motion is primarily made of cooperative twisting of helix α4 and helix α5. This motion covers the same region as the first essential motion, further expands to the helix α5, and, interestingly, also to the loop before helix α8, which is connected with the flexible region around helix α4 by the β-bridge. The maximum amplitude of the second essential motion is localized around the residues Ile138, Pro212, and Ala214 participating in the β-bridge. The β-bridge is formed by two backbone hydrogen bonds: Ile138-NH. . .O-Pro212 and Ala214-NH. . .O-Ile138. The first hydrogen bond remains for 98.6% of the trajectory with a mean distance of 3.2 Å and a mean angle of 12.9°. The second hydrogen bond remains for 99.5% of the trajectory with a mean distance of 2.9 Å and a mean angle of 12.8°. There is one additional short living H-bond formed between Ala 214-NH. . .O-Pro 212 occupying 7.7% of the trajectory with a mean distance of 3.7 Å and a mean angle of 51.6°.
The region participating in the first concerted motion of DhaA covers the random coil before helix α4, the first half of helix α4, and the loop before helix α8 connected with the random coil before helix α4 by the β-bridge (Fig. 2 ▶). The first essential motion of DhaA is very similar to the first essential motion of LinB but has smaller amplitude and is more localized at helix α4 and random coil before helix α4, whereas helix α5′ and helix α5 remain static. Essentially, the same region is also involved in the second concerted motion of DhaA. Both essential motions are partly transferred to the loop before helix α8 because of the β-bridge. The β-bridge is formed by two backbone hydrogen bonds: Ile135-NH. . .O-Pro210 and Ala212-NH. . .O-Ile135. The first hydrogen bond remains for 98.2% of the trajectory with a mean distance of 3.2 Å and a mean angle of 20.3°. The second hydrogen bond remains for 99.5% of the trajectory with a mean distance of 3.0 Å and a mean angle of 18.1°. The maximum amplitude of the essential motions occurs around the residues Asp139, Glu140, and Glu143.
Dynamics of the secondary structure elements
Most of the secondary structure elements in all three studied proteins remained intact during the simulation. Generally, the β-strands were better defined during the simulation than the α-helices. This observation is consistent with the fact that the central β-sheet forms the core of the haloalkane dehalogenases. Another general observation is that the least stable α-helices are always located in the cap domains.
The helix α4′ and, to a smaller extent, the first half of the helix α7, are the only unstable secondary elements of DhlA (Fig. 3 ▶). The C-terminal end of the helix α4′ gets extended at several points of the trajectory. More importantly, the entire secondary structure of this helix is disrupted in times between 490 ps and 660 ps.
Fig. 3.
Development of the secondary structure elements during the simulation. α-helices are in black and β-strands are in gray. The secondary structure elements were assigned using the program dssp as described in the Materials and Methods section. The labels of secondary structure elements are indicated on the right side of each graph.
The helix α4, helix α5′, and helix α5 are the most unstable secondary structure elements of LinB (Fig. 3 ▶). Two parts of the helix α5 can be distinguished in LinB and DhaA based on its structure and dynamics. The N-terminal part of helix α5 of LinB is more stable compared with the C-terminal part, which is very unstable balancing between an α-helix (i.e., Pauling-Corey 3.613-helix) and a 310-helix.
The helix α4 and helix α5 are the most unstable secondary structure elements of DhaA (Fig. 3 ▶). Both parts of helix α5, defined in the previous paragraph, are flexible. The C-terminal part is balancing between an α-helix and a 310-helix.
Dynamics of the amino acid residues
Atomic temperature factor (B-factor) is a measure of thermal mobility and disorder of the individual atom in the protein molecule. B-factor values derived from the crystallographic analysis are compared with the values calculated from the simulation (Fig. 4 ▶). The B-factor values obtained here via simulation are on average larger than those from X-ray (Karplus and McCammon 1983). The regions with higher B-factors from the crystallographic analysis generally show higher B-factors from the molecular dynamic simulations. These flexible regions mainly correspond to the loops and random coils.
Fig. 4.
Comparison of the temperature factors (in Å2) obtained from the crystallographic analysis (thin line) and from the molecular dynamic simulation (thick line). Secondary structure elements are indicated on the x-axis (α-helices as the black bars, β-strands as the gray bars). The temperature factors for the crystal structures were extracted from PDB files (Verschueren et al. 1993b;Newman et al., 1999; Marek et al., 2000).
The catalytic residues of the entire family of haloalkane dehalogenases are organized in a catalytic triad, which is composed of a nucleophile, an acid, and a base. The nucleophile Asp124 of DhlA is significantly more rigid (B-factor 8.9) compared with the catalytic acid Asp260 (12.6) and the base His289 (11.6). A halogen group of the substrates of haloalkane dehalogenases and the halide ion formed during the reaction are stabilized by Trp125, Phe172, and Trp175 in DhlA. Trp125 is rigid (9.8) but Phe172 (16.4) and, especially, Trp175 (21.3) are flexible. The high flexibility of Trp175 is unexpected considering its important role for the catalysis. According to B-factors, the most flexible helices are helix α4, helix α5, and helix α8, whereas the most flexible region of the entire DhlA structure is the loop between helix α4′ and helix α4 (Fig. 4 ▶). The most flexible residues of DhlA are located in this region: Asp170 (40.0), Ala169 (38.6), Gln167 (34.9), and Thr166 (28.1). The flexibility of the region is dependent on the presence or absence of the salt bridge between Asp170 and Lys261 (data not shown). The other highly flexible residues of DhlA are Glu47 (38.7) and Asp45 (31.8), located on the loop between strand β2 and strand β3; Arg193 (33.8) and Trp194 (29.7) from the C-terminal part of helix α4; and Asn246 (32.1) and Gln245 (31.3) from the C-terminal part of helix α8.
Two of three catalytic triad residues of LinB, Asp108 (9.7), and Glu132 (9.5) are among the most rigid residues in the protein. The third residue of the triad His282 (11.5) can still be considered as rigid compared with the rest of the protein. Two halide-stabilizing residues, Asn38 (12.7) and Trp109 (11.75), are relatively rigid. The other two residues that are in direct contact with the halide ion bound to the active site, Phe151 (27.4) and Phe169 (20.6), are highly flexible. Phe151 is one of the most flexible residues of this protein. The flexible helices of LinB are helix α4, helix α5′, and helix α6 (Fig. 4 ▶). The most flexible region of the entire LinB is the random coil before helix α4 with the extremes in Gln146 (31.9), Glu139 (29.6), Glu145 (27.3), and Asp147 (24.8); and the loop between helix α4 and helix α5′ with highly flexible Gln165 (30.5), Glu161 (30.0), Gln157 (28.4), Phe151 (27.4), Gln152 (25.1), and Asp166 (23.8).
All three catalytic triad residues of DhaA—Asp106 (9.3), Glu130 (8.69), and His272 (9.67)—are extremely rigid. Two halide-stabilizing residues, Asn41 (11.71) and Trp107 (10.79), are similarly rigid as in LinB, whereas two other residues in contact with the halide ion, Phe149 (12.39) and Phe168 (13.52), are considerably more rigid than analogous residues in LinB. The two most flexible helices of DhaA, according to B-factors, are helix α4 and helix α6 (Fig. 4 ▶). The most flexible region of the entire DhaA is the random coil before helix α4 with extremes in Glu140 (26.8), Glu143 (26.0), Glu147 (24.8), Asp139 (24.4), and Thr136 (22.8). The two most flexible residues of helix α6 are His188 (24.7) and Asp183 (23.5).
Dynamics of the solvent molecules
Mobility of the active-site water molecules and their exchange with the bulk solvent during the simulation was analyzed throughout the trajectory. The catalytic water molecules were significantly less mobile than the rest of the active-site water molecules in all of the studied proteins (data not shown). Catalytic waters remained in their crystal positions during the entire simulations. This is the only feature in common with all three dehalogenases because the exchange of water molecules between the active sites and bulk solvent varied significantly among the proteins.
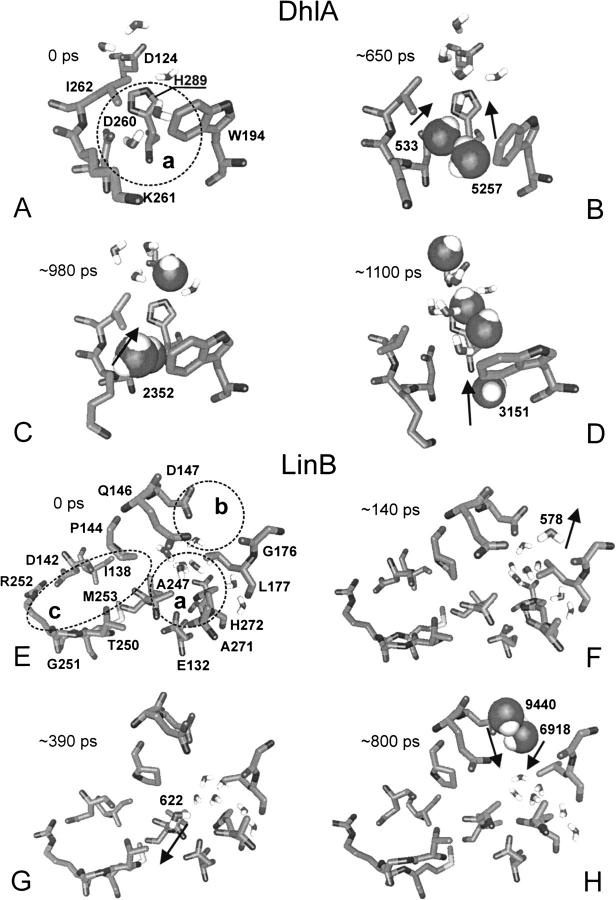
There were five water molecules in the active site of DhlA at the beginning of the simulation (Fig. 5A ▶). Four additional water molecules entered the active site via the lower tunnel (residues Trp194, Asp260, Lys261, Leu262, His289, and Phe290; the assignment of the entrance tunnels used in this study is based on the location of the corresponding upper tunnel, lower tunnel, and a slot in LinB structure; Fig. 6 ▶), whereas no molecule left the active site during the simulation. The penetration of the water molecules from a bulk solvent to the active site occurred as follows: the water molecule 533 approached the entrance of the lower tunnel at ∼40 ps and another water molecule 5257 came to the entrance of the same tunnel at ∼570 ps. Both water molecules entered the active site between Lys261 and Trp194 at ∼650 ps (Fig. 5B ▶). The water molecule 2352 entered the active site through the lower tunnel at ∼980 ps (Fig. 5C ▶). The water molecule 3151 entered the active site at the end of the simulation (Fig. 5D ▶).
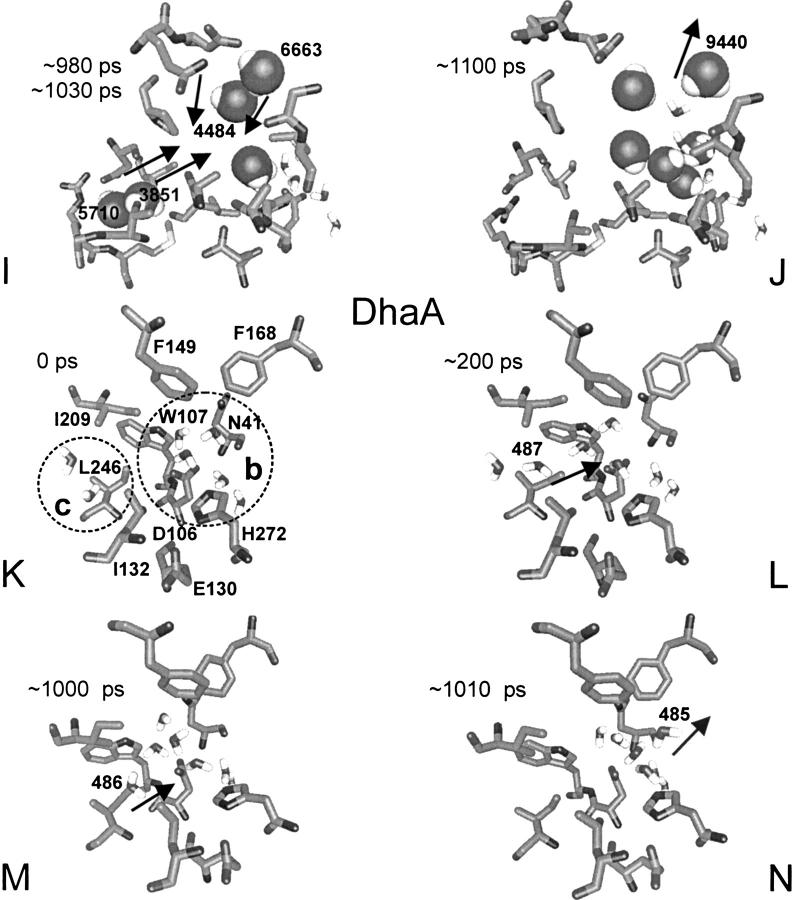
Fig. 5.
Exchange of the water molecules between the bulk solvent and the active sites of DhlA (A–D), LinB (E–J), and DhaA (K–N) along the trajectory. Selected active-site and tunnel residues are shown in stick. The active site and the slot water molecules are in stick, and the bulk solvent molecules entering the active site are in ball. The times of exchange (in picoseconds) are indicated in every snapshot. Direction of the water molecules exchange between the active site, a bulk solvent, and the slot, respectively, is indicated by the arrow. Circle (a) denotes the lower tunnel, circle (b) denotes the upper tunnel, and circle (c) denotes the slot. The tunnels are named according to their location within LinB structure (Fig. 6 ▶).
Fig. 6.
Stereo view of ribbon representation of the LinB structure; denoted are the lower tunnel (a), the upper tunnel (b), and the slot (c). Selected residues separating the tunnels are labeled.
Frequent exchange of the water molecules between the bulk solvent and the active site of LinB was observed. Six water molecules from a bulk solvent entered the enzyme active site, and three molecules left the active site during the simulation. There were eight water molecules in the active site of LinB at the beginning of the simulation (Fig. 5E ▶). The active-site water molecule 578 left the active site through the upper tunnel (residues Gln146, Asp147, Gly176, and Leu177) at ∼140 ps (Fig. 5F ▶). The water molecule 622 left the active site through the lower tunnel (residues Gln146, Leu177, Ala247, Ala271, and His272) at ∼390 ps (Fig. 5G ▶). The two bulk water molecules 6918 and 9440 entered the active site through the upper tunnel at ∼800 ps (Fig. 5H ▶), followed by the two additional water molecules 6663 and 4484 at ∼980 ps (Fig. 5I ▶). The two water molecules 3851 and 5710 entered the active site through the slot (residues Ile138, Asp142, Pro144, Ala247, Leu248, Thr250, Gly251, and Arg252) at ∼1030 ps (Fig. 5I ▶). The water molecule 9440 left the active site through the upper tunnel at the end of the simulation (Fig. 5J ▶).
There were five water molecules present in the active site of DhaA at the beginning of the simulation. Two additional water molecules were in the slot adjacent to the active site (Fig. 5K ▶). The water molecule 487 from the slot entered the active site at ∼200 ps (Fig. 5L ▶). The second water molecule 486 from the slot entered the active site at ∼1000 ps (Fig. 5M ▶). The water molecule 485 left the active site through the upper tunnel (residues Phe144, Ala145, Phe168, Val172, Lys175, and Cys176) at ∼1010 ps soon after the entrance of the second water molecule (Fig. 5N ▶). No water from a bulk solvent entered the active site during the entire simulation.
Discussion
The motions observed during 1-ns simulations of the three haloalkane dehalogenases were related to their structural and catalytic properties, keeping in mind that these enzymes perform a catalytic cycle on a second time scale and therefore the catalytically relevant motions occurring with low frequency or very slowly could not be observed during these simulations. The main observations of this study and their biochemical relevance are listed here and discussed in more detail in the following text. (1) The main domains of the haloalkane dehalogenases are rigid. They provide a skeleton for hanging on the catalytic residues. Analogous domains occur in many proteins of the α/β-hydrolase fold and should also possess a high level of rigidity. (2) The cap domains are much more flexible than the main domains and display concerted motions. The localization and character of concerted motions differ among haloalkane dehalogenases and may influence their substrate specificity. (3) The most flexible part of the haloalkane dehalogenases is the random coil interconnecting two domains. This flexibility is the consequence of the modular composition of haloalkane dehalogenases and therefore may be found in other proteins as well. Haloalkane dehalogenases use a salt bridge and a β-bridge, respectively, for stabilization of this highly flexible region. (4) The nucleophile and the catalytic base show the same level of flexibility in different haloalkane dehalogenases, the former being one of the most rigid residues in these proteins. High rigidity of the nucleophile and the base is essential for the catalysis and may be extrapolated to other members of the α/β-hydrolase fold. Variable flexibility of the catalytic acid is the consequence, or the purpose, of its migration within the protein structure and may influence the rate of the second catalytic step. (5) The catalytic water of haloalkane dehalogenases is significantly less mobile than other water molecules in the active site. Good stabilization and positioning of the catalytic water molecule are essential for the reaction and should also be required by other proteins using the water during their catalytic cycle. (6) Exchange of water molecules between the active-site cavity and bulk solvent differs among dehalogenases as the consequence of the different mobility of the cap domains and different number of the entrance tunnels.
No functionally relevant motions were detected in the main domains by the essential dynamics analysis. The analysis of B-factors showed that the β-pleated sheet is the most rigid part of a protein and the analysis of secondary elements confirmed the high stability of all β-strands along trajectories. These observations are in line with the primary role of the main domain to provide the skeleton for hanging on the catalytic residues in a spatial arrangement suitable for catalysis (Ollis et al. 1992). It can also be expected that other members of the α/β-hydrolase fold will show high rigidity of their main domains. The knowledge of rigid regions of these proteins is important for the experiments attempting to apply the phage display system for directed evolution.
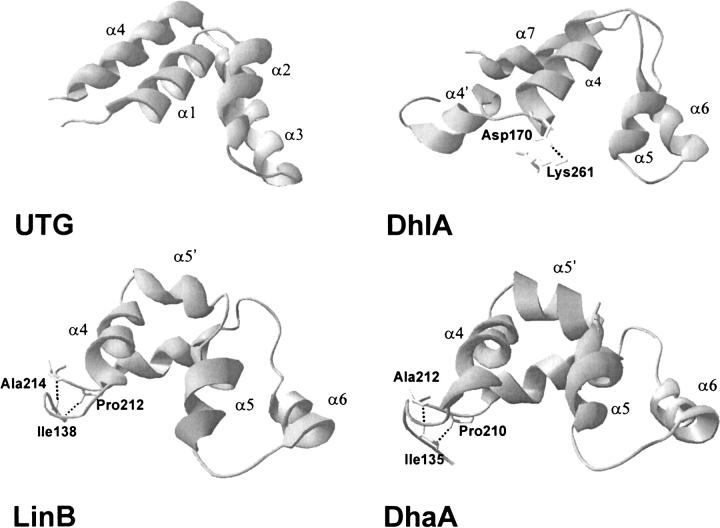
Unlike the main domains, the cap domains of all three dehalogenases displayed concerted motions. Localization and character of these motions was, however, different in different enzymes. Similarly, the architecture of the cap domain of DhlA is different from that in LinB and DhaA (Fig. 7 ▶), and the substrate specificity of DhlA is significantly different from the specificity of LinB and DhaA (Damborsky et al. 1997b; Damborsky et al. 2001). The essential motions of DhlA are spread over the entire cap domain. All but helix α4′ in the cap domain of DhlA remained intact during the simulation, indicating that essential motions are caused by displacement of the secondary elements relative to each other. The lower stability of the helix α4′ can be related to the arrangement of domains in DhlA: the secondary elements before helix α4′ belong to the α/β-hydrolase fold domain, whereas the secondary elements after this helix belong to the UTG-fold domain. Helix α4′ creates the linkage between these two domains and shows partially distorted helical geometry. We propose that random coil was present in that region, as in LinB and DhaA, during the early stage of evolution of DhlA and imperfect helix was created by insertion to improve the stability and packing of the structure. Two direct repeats are present in this region as noted by Pries and coworkers (Pries et al. 1994). The large conformational change responsible for the release of the halide ion from the enzyme active site has been proposed in the helix α4′-loop-helix α4 region (Schanstra and Janssen 1996; Krooshof et al. 1999). Although this conformational change occurs on a millisecond time scale, the highest mobility of this region during the nanosecond simulation indicates that this part of the protein has a predisposition for occurrence of such a conformational change. The salt bridge between Asp170 and Lys261, connecting highly flexible helix α4′-loop-helix α4 with rigid main domain, appears to be an important structural element for fine-tuning of the flexibility in this region. There must be a trade-off between the rigidity needed for stabilization of dehalogenation reaction by the active-site residues Phe172 and Trp175 located in the α4-helix and the flexibility needed for release of halide ion from the active site after the reaction. Certain flexibility of the entire cap domain can also be important for binding and catalysis of structurally different substrates that have to penetrate into the small active site via narrow tunnel. These observations correspond well with the earlier proposal of Pries at al. (1994) and Kmunicek et al. (2001) that the cap domain of DhlA determines its substrate specificity. Pries et al. reported six different in vivo mutants of DhlA (including Asp170His) with improved catalytic efficiency with 1-chlorohexane. All mutations occurred in the N-terminal part of the cap domain and the investigators proposed that improved binding of this large substrate is attributable to larger or more flexible active-site cavity (Pries at al. 1994). The active sites of LinB and DhaA are less buried and their entrance tunnels are wider compared with DhlA (Damborsky and Koca 1999; Marek et al. 2000). Similarly, the essential motions of LinB and DhaA are localized to smaller regions of the cap domain. The random coil before helix α4 appears to be the most flexible region of LinB and DhaA. This region forms the linkage between the α/β-hydrolase fold domain and UTG-fold domain. Different dehalogenases apparently stabilize this interdomain region by different structural elements: DhlA evolved the α4′-helix and the salt bridge, whereas LinB and DhaA evolved the β-bridge (Fig. 7 ▶). As a consequence of this β-bridge, essential motions in LinB and DhaA are transferred to the loop before helix α8 of the main domain. The major difference between the essential motions of LinB and DhaA is in the region of helix α5′ and helix α5. The helix α5′ of LinB stretches and snubs along its main axis because it is perpendicular to both helix α4 and helix α5 and possesses enough space in this direction (Fig. 7 ▶). In DhaA, the helix α5′ is contained between helix α4 and helix α5, resulting in a more compact structure (Fig. 7 ▶). The different arrangement of helix α4, helix α5′, and helix α5 in LinB and DhaA is attributable to the different amino acid composition and structure of the loop connecting helix α4 with helix α5′. Ala156 located in this loop of DhaA has angles φ and ξ in an unfavorable region of the Ramachandran plot (Newman et al. 1999) and remains in this region along the entire trajectory. In contrast, LinB carries deletion in a position equivalent to 157 of DhaA and all amino acids of this loop have φ and ξ angles in favorable regions of the Ramachandran plot. In evolutionary terms, LinB appears to be better adapted to the structural requirements possessed on the loops in cap domain than DhaA. Functionally, the difference in composition and dynamics of the cap domains may influence the ability and kinetics of binding of different substrates to the active sites through the tunnels located in these regions. LinB shows higher activity with β-substituted haloalkanes compared with DhaA (Damborsky et al. 2001). Higher flexibility of cap domain in LinB may be important for proper positioning of these substrates for the SN2 dehalogenation reaction. The different dynamics of the cap domains of LinB and DhaA are also reflected by the very different exchange of water molecules between their active sites and bulk solvent (discussed below).
Fig. 7.
Ribbon representation of uteroglobin monomer and the cap domains of haloalkane dehalogenases. UTG stands for uteroglobin (PDB ID 1UTG). The residues participating on the salt bridge (DhlA) and the β-bridges (LinB, DhaA) are labeled.
There are several amino acid residues in the active sites of haloalkane dehalogenases that are known to be essential (catalytic triad) or very important (halide-stabilizing residues) for an enzymatic catalysis. Mutations in the residues of the catalytic triad result in loss of activity (Pries et al. 1995a; Pries et al. 1995b; Hynkova et al. 1999), whereas mutations in the halide-stabilizing residues result in significantly reduced activity (Kennes et al. 1995; Schindler et al. 1999). The residues of the catalytic triad are directly involved in the dehalogenation reaction and their lower flexibility is expected to be favorable for the catalysis. The same level of flexibility was found for the nucleophile and the base in all three dehalogenases, the former being one of the most rigid residues in haloalkane dehalogenases; however, a significant difference was observed for the catalytic acid, which is significantly more flexible in DhlA than in LinB and DhaA. The higher flexibility of the catalytic acid of DhlA was also observed in the molecular dynamics study of Arnold and Ornstein (Arnold and Ornstein 1997). The catalytic acid is located after strand β6 in LinB and DhaA, whereas it is after strand β7 in DhlA (Krooshof et al. 1997; Hynkova et al. 1999). Requirements for proper flexibility of the catalytic acid could be one of the selection factors responsible for the migration of the catalytic acid in haloalkane dehalogenases (Krooshof et al. 1997) and some other α/β-hydrolases (Schrag et al. 1992). Halide-stabilizing residues of haloalkane dehalogenases are involved in several steps of the catalytic cycle: (1) interact with the halogen atom of the substrate on its binding to a Michaelis-Menten complex (Verschueren et al. 1993a), (2) stabilize transition state of the first reaction step (Damborsky et al. 1997a; Lightstone et al. 1997), and (3) stabilize halide ion released from the substrate during the first reaction step (Verschueren et al. 1993a; Damborsky et al. 1997a). Dynamic behavior of these residues may influence any of these catalytic steps but may also impact the specificity of dehalogenases toward chlorinated, brominated, and iodinated substrates. The halide-stabilizing tryptophan positioned adjacent to the nucleophile (Trp125, Trp109 and Trp107 in DhlA, LinB, and DhaA, respectively) is conserved in all three proteins. This residue shows comparable flexibility in different dehalogenases. The second halide-stabilizing residue (Trp175, Asn38, and Asn41, respectively) shows significantly higher flexibility in DhlA compared with LinB and DhaA. High flexibility of Trp175 may be required for destabilization of the electrostatic interactions between the partially positively charged nitrogen atom of Trp175 and negatively charged halide ion during a release of the halide ion from the buried active site. According to Lightstone and coworkers, Trp175 primarily stabilizes the transition state of the SN2 reaction step but is less involved in the stabilization of the substrate in the Michaelis-Menten complex (Lightstone et al. 1998). Weaker halide stabilization in the active site of DhaA and LinB, proposed from quantum-mechanic calculations (Damborsky et al. 1997a) and kinetic studies (Schindler et al. 1999), indicates that flexibility of Asn38 and Asn41, respectively, is not needed for efficient catalysis.
The second reaction step of the haloalkane dehalogenases is the hydrolysis of alkyl-enzyme intermediate by a water molecule (Janssen et al. 1985). Analysis of mobility of the water molecules occurring in the active site during the simulation revealed that catalytic water molecules show the lowest mobility of all molecules present. This is attributable to the hydrogen bonds between the protein molecule and the catalytic water and to the hole made of the residues surrounding the catalytic water in the enzyme active site. Apparently, hydrogen bonding network and the geometry of the active site is adjusted to bind catalytic waters in a position optimal for the reaction. Exchange of water molecules between the enzyme active site and the bulk solvent was analyzed. Four water molecules entered and no molecule left the active site of DhlA during the simulation. Nine water molecules in total accumulated in the active site of this protein because five crystallographically resolved molecules were in the active site from the beginning of the simulation. This is an unexpected observation considering the small size, buried position, and high hydrophobicity of the active site of this protein. DhlA can apparently relax its structure to enlarge its active site. Loops of entire cap domain and especially helix α4 are involved in this relaxation, as seen from the essential dynamics and analysis of secondary structure elements along the trajectory.
Six water molecules entered and five molecules left the active site of LinB during the simulation. Very frequent exchange of water molecules in LinB can be related to a number of tunnels leading to the active site from the bulk solvent but also to the mobility of the cap domain of this protein. There are at least three routes, that is, upper tunnel, lower tunnel, and slot (Fig. 6 ▶), that can be explored by the water molecules to access and leave the enzyme active site. The secondary structure elements in the cap domain of LinB show the highest mobility. The ability of LinB to adjust the size of its tunnels and the active site can be essential for binding and catalysis of large substrates such as bromocycloheptane and bromomethylcyclohexane (Damborsky et al. 2001) or its natural substrate 1,3,4,6-tetrachloro-1,4-cyclohexadiene (Nagata et al. 1999). Two water molecules entered the active site of DhaA from the slot and one water molecule left the active site immediately after the entrance of the second molecule. The active site of DhaA apparently cannot accept more than six water molecules at a time. This is consistent with the lowest level of essential motions in DhaA compared with LinB and DhlA. Low frequency of the exchange of water molecules between the solvent and the active site may also be related to the lack of the lower tunnel. Unlike LinB, DhaA does not have to catalyze dehalogenation of large cyclic compounds under physiological conditions. It is also consistent with more restricted substrate specificity of DhaA toward larger β-substituted compounds in comparison with LinB (Damborsky et al. 2001).
Materials and methods
Protein structures
Molecular dynamics simulations were performed with the structure of haloalkane dehalogenases from Xanthobacter autotrophicus GJ10 (DhlA), Sphingomonas paucimobilis UT26 (LinB), and Rhodococcus sp. (DhaA) using the SANDER module of AMBER 5.0 (Case et al. 1997) and force field of Cornell et al. (Cornell et al. 1995). The starting geometries were prepared from the X-ray structures obtained from the Protein Data Bank—PDB access codes 2HAD (Verschueren et al. 1993b), 1CV2 (Marek et al. 2000), and 1CQW (Newman et al. 1999). The X-ray structures were prepared for molecular dynamic simulations as follows. The hydrogen atoms were added to the X-ray structures using the program WHATIF 5.0 (Vriend 1990). All heavy atoms were restrained and the molecule was minimized in vacuum using the program SANDER (1500 optimization steps). The all-atom model was neutralized by adding the required amount of counter ions using the program GRID (Goodford 1985). The enzyme with crystal water was immersed in a rectangular water box. The layer of water molecules was 10 Å. The initial size of the box was equal to 73 Å × 75 Å × 65 Å for DhlA and it contained 9139 TIP3P water molecules and 17 Na+ counter ions; 82 Å × 64 Å × 64 Å for LinB and it contained 9439 TIP3P water molecules and 12 Na+ counter ions; and 81 Å × 68 Å × 70 Å for DhaA and it contained 11 579 TIP3P water molecules and 18 Na+ counter ions.
Minimization
The protein-solvent system was optimized before the simulation. First, the protein was frozen and the solvent molecules with counter ions were allowed to move during a 1000-step minimization of steepest descent and a 2-ps molecular dynamics run. Second, the side chains were allowed to relax by several subsequent minimizations of steepest descent during which decreasing force constants were put on the backbone atoms. After full relaxation, the system was slowly heated to 250 K in 10 ps and then to 300 K in 40 ps.
Simulation
Two femtosecond time step and the Particle Mesh Ewald (PME) method (Essmann et al. 1995) were used in all simulations. The simulations were initiated under the periodic boundary condition in the NpT ensemble at 300 K with Berendsen temperature coupling (Berendsen et al. 1984). The SHAKE algorithm (Ryckaert et al. 1977) was applied to fix all bonds containing a hydrogen atom, and the nonbond pair list was updated every 10 steps. A 9.0 Å cutoff was applied to Lennard-Jones interactions. The ensemble with constant number of particles, volume, and temperature (NVT) was used after the density became balanced. The NVT part of the simulations was 1100 ps long and it was considered to be a production part. Coordinates were written to trajectory files after each picosecond.
Data analysis
The results from simulations were analyzed using the Carnal and Ptraj modules of the AMBER 5.0 package. The B-factors were calculated as described in the literature (Resat and Mezei 1996) using the equation Bi = (8π2/3) • Ri2, where Bi and Ri are the B-factor and RMSD of the atom i, respectively. The B-factor of the residue was obtained by averaging over temperature factors of all atoms in the residue. Obtained B-factors were compared with the values from X-ray analysis. The trajectories were further analyzed for essential motions (Amadei et al. 1993) using the programs g_covar and g_anaeig of the GROMACS 2.0 program package (van der Spoel et al. 1999). The projected eigenvectors were visualized using the MOLMOL 2K.1 software (Koradi et al. 1996). The program do_dssp of GROMACS 2.0 was used for analysis of changes in the secondary elements during the simulation (Kabsch and Sander 1983). The solvent-accessible surface was computed using the do_dssp and the program NACCESS 2.1.1 (Lee and Richards 1971). The molecular dynamic trajectories were visualized by using the gOpenMol 2.0 program package (Laaksonen 2001).
Acknowledgments
We thank Dr. Rebecca Wade (EMBL, Germany) for valuable advice on simulations at the early stage of the project and Drs. Ton Linssen (Utrecht University, The Netherlands) and Jaap Flohil (Technical University Delft, The Netherlands) for their help with setting up the GROMACS package. The Czech Academic Supercomputing Centers are acknowledged for providing computer time and technical support. This work was supported by the grants MSM 153100008, 1562/2001, and LN00A016 from the Ministry of Education of the Czech Republic.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps3830102
References
- Amadei, A., Linssen, A.B.M., and Berenden, H.J.C. 1993. Essential dynamics of protein. Proteins: Struct. Funct. Gen. 17 412–425. [DOI] [PubMed] [Google Scholar]
- Arnold, G.E. and Ornstein, R.L. 1997. Molecular dynamics study of haloalkane dehalogenase; implications for solvent channel access to the active site. In Biomacromolecules: From 3-D to applications (ed. R.L. Ornstein), pp. 215–229. Battelle Press, Columbus, Ohio.
- Berendsen, H.J.C., Postma, J.P.M., van Gunsteren, W.F., DiNola, A., and Haak, J.R. 1984. Molecular-dynamics with coupling to an external bath. J. Comp. Phys. 81 3684–3690. [Google Scholar]
- Case, D.A., Pearlman, D.A., Caldwell, J.W., Cheatham III, T.E., Ross, W.S., Simmerling, C.L., Darden, T.A., Merz, K.M., Stanton, R.V., Cheng, A.L., et al. 1997. AMBER 5.0. University of California, San Francisco.
- Copley, S.D. 1998. Microbial dehalogenases: Enzymes recruited to convert xenobiotic substrates. Curr. Opin. Chem. Biol. 2 613–617. [DOI] [PubMed] [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117 5179–5197. [Google Scholar]
- Damborsky, J. and Koca, J. 1999. Analysis of the reaction mechanism and substrate specificity of haloalkane dehalogenases by sequential and structural comparisons. Protein Eng. 12 989–998. [DOI] [PubMed] [Google Scholar]
- Damborsky, J., Kuty, M., Nemec, M., and Koca, J. 1997a. A molecular modeling study of the catalytic mechanism of haloalkane dehalogenase: 1. Quantum chemical study of the first reaction step. J. Chem. Inf. Comp. Sci. 37 562–568. [Google Scholar]
- Damborsky, J., Nyandoroh, M.G., Nemec, M., Holoubek, I., Bull, A.T., and Hardman, D.J. 1997b. Some biochemical properties and classification of a range of bacterial haloalkane dehalogenases. Biotech. Appl. Biochem. 26 19–25. [PubMed] [Google Scholar]
- Damborsky, J., Rorije, E., Jesenska, A., Nagata, Y., Klopman, G., and Peijnenburg, W.J.G.M. 2001. Structure-specificity relationships for haloalkane dehalogenases. Environ. Toxicol. Chem. 20 2681–2689. [PubMed] [Google Scholar]
- Essmann, U., Perera, L., Berkowitz, M.L., Darden, T.A., Lee, H., and Pedersen, L.G. 1995. A Smooth Particle Mesh Ewald method. J. Chem. Phys. 103 8577–8593. [Google Scholar]
- Goodford, P.J. 1985. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 28 849–857. [DOI] [PubMed] [Google Scholar]
- Hynkova, K., Nagata, Y., Takagi, M., and Damborsky, J. 1999. Identification of the catalytic triad in the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. FEBS Lett. 446 177–181. [DOI] [PubMed] [Google Scholar]
- Janssen, D.B., Pries, F., and Van der Ploeg, J.R. 1994. Genetics and biochemistry of dehalogenating enzymes. Annu. Rev. Microbiol. 48 163–191. [DOI] [PubMed] [Google Scholar]
- Janssen, D.B., Scheper, A., Dijkhuizen, L., and Witholt, B. 1985. Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl. Environ. Microbiol. 49 673–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Karplus, M. and McCammon, J.A. 1983. Dynamics of proteins: Elements and functions. Ann. Rev. Biochem. 53 263–300. [DOI] [PubMed] [Google Scholar]
- Kennes, C., Pries, F., Krooshof, G.H., Bokma, E., Kingma, J., and Janssen, D.B. 1995. Replacement of tryptophan residues in haloalkane dehalogenase reduces halide binding and catalytic activity. Eur. J. Biochem. 228 403–407. [PubMed] [Google Scholar]
- Kmunicek, J., Luengo, S., Gago, F., Ortiz, A.R., Wade, R.C., and Damborsky, J. 2001. Comparative binding energy analysis of the substrate specificity of haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. Biochemistry 40 8905–8917. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wurthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- Krooshof, G.H., Floris, R., Tepper, A., and Janssen, D.B. 1999. Thermodynamic analysis of halide binding to haloalkane dehalogenase suggests the occurrence of large conformational changes. Protein Sci. 8 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krooshof, G.H., Kwant, E.M., Damborsky, J., Koca, J., and Janssen, D.B. 1997. Repositioning the catalytic triad acid of haloalkane dehalogenase: Effects on activity and kinetics. Biochemistry 36 9571–9580. [DOI] [PubMed] [Google Scholar]
- Laaksonen, L. 2001. gOpenMol 2.1. Espoo, Finland.
- Lee, B. and Richards, F.M. 1971. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 55 379–400. [DOI] [PubMed] [Google Scholar]
- Lightstone, F.C., Zheng, Y.J., and Bruice, T.C. 1998. Molecular dynamics simulations of ground and transition states for the S(N)2 displacement of Cl- from 1,2-dichloroethane at the active site of Xanthobacter autotrophicus haloalkane dehalogenase. J. Am. Chem. Soc. 120 5611–5621. [Google Scholar]
- Lightstone, F.C., Zheng, Y.-J., Maulitz, A.H., and Bruice, T.C. 1997. Non-enzymatic and enzymatic hydrolysis of alkyl halides: A haloalkane dehalogenation enzyme evolved to stabilize the gas-phase transition state of an SN2 displacement reaction. Proc. Natl. Acad. Sci. 94 8417–8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek, J., Vevodova, J., Kuta-Smatanova, I., Nagata, Y., Svensson, L.A., Newman, J., Takagi, M., and Damborsky, J. 2000. Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26. Biochemistry 39 14082–14086. [DOI] [PubMed] [Google Scholar]
- Nagata, Y., Miyauchi, K., Damborsky, J., Manova, K., Ansorgova, A., and Takagi, M. 1997. Purification and characterization of haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl. Environ. Microbiol. 63 3707–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, Y., Miyauchi, K., and Takagi, M. 1999. Complete analysis of genes and enzymes for γ-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J. Ind. Microbiol. Biotech. 23 380–390. [DOI] [PubMed] [Google Scholar]
- Nardini, M. and Dijsktra, B.W. 1999. α/β Hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 9 732–737. [DOI] [PubMed] [Google Scholar]
- Newman, J., Peat, T.S., Richard, R., Kan, L., Swanson, P.E., Affholter, J.A., Holmes, I.H., Schindler, J.F., Unkefer, C.J., and Terwilliger, T.C. 1999. Haloalkane dehalogenase: Structure of a Rhodococcus enzyme. Biochemistry 38 16105–16114. [DOI] [PubMed] [Google Scholar]
- Ollis, D.L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S.M., Harel, M., Remington, S.J., Silman, I., Schrag, J., et al. 1992. The α/β hydrolase fold. Protein Eng. 5 197–211. [DOI] [PubMed] [Google Scholar]
- Poelarends, G., Zandstra, M., Bosma, T., Kulakov, L.A., Larkin, M.J., Marchesi, J.R., Weightman, A.J., and Janssen, D.B. 2000. Haloalkane-utilizing Rhodococcus strains isolated from geographically distinct locations possess a highly conserved gene cluster encoding haloalkane catabolism. J. Bacteriol. 182 2725–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries, F., Kingma, J., and Janssen, D.B. 1995a. Activation of an Asp-124–Asn mutant of haloalkane dehalogenase by hydrolytic deamidation of asparagine. FEBS Lett. 358 171–174. [DOI] [PubMed] [Google Scholar]
- Pries, F., Kingma, J., Krooshof, G.H., Jeronimus-Stratingh, C.M., Bruins, A.P., and Janssen, D.B. 1995b. Histidine 289 is essential for hydrolysis of the alkyl-enzyme intermediate of haloalkane dehalogenase. J. Biol. Chem. 270 10405–10411. [DOI] [PubMed] [Google Scholar]
- Pries, F., Van den Wijngaard, A.J., Bos, R., Pentenga, M., and Janssen, D.B. 1994. The role of spontaneous cap domain mutations in haloalkane dehalogenase specificity and evolution. J. Biol. Chem. 269 17490–17494. [PubMed] [Google Scholar]
- Resat, H. and Mezei, M. 1996. Grand Canonical Ensemble Monte Carlo simulation of the dCpG/proflavine crystal hydrate. Biophys. J. 71 1179–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, R.B. and Sternberg, M.J.E. 1997. Two new examples of protein structural similarities within the structure-function twilight zone. Protein Eng. 10 333–338. [DOI] [PubMed] [Google Scholar]
- Ryckaert, J.P., Ciccotti, G., and Berendsen, H.C. 1977. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Phys. 23 327–341. [Google Scholar]
- Schanstra, J.P. and Janssen, D.B. 1996. Kinetics of halide release of haloalkane dehalogenase: Evidence for a slow conformational change. Biochemistry 35 5624–5632. [DOI] [PubMed] [Google Scholar]
- Schindler, J.F., Naranjo, P.A., Honaberger, D.A., Chang, C.-H., Brainard, J.R., Vanderberg, L.A., and Unkefer, C.J. 1999. Haloalkane dehalogenases: Steady-state kinetics and halide inhibition. Biochemistry 38 5772–5778. [DOI] [PubMed] [Google Scholar]
- Schrag, J.D., Winkler, F.K., and Cygler, M. 1992. Pancreatic lipases: Evolutionary intermediates in a positional change of catalytic carboxylates? J. Biol. Chem. 267 4300–4303. [PubMed] [Google Scholar]
- Slater, J.H., Bull, A.T., and Hardman, D.J. 1995. Microbial dehalogenation. Biodegradation 6 181–189. [Google Scholar]
- van der Spoel, D., van Buuren, A.R., Apol, E., Maulenhoff, P.J., Tieleman, P.D., Sijbers, A.L.T.M., Hess, B., Feenstra, K.A., Lindhal, E., Drunen, R.V., et al. 1999. Gromacs user manual version 2.0. Groningen, The Netherlands.
- Verschueren, K.H.G., Franken, S.M., Rozeboom, H.J., Kalk, K.H., and Dijkstra, B.W. 1993a. Non-covalent binding of the heavy atom compound [Au(CN)2]- at the halide binding site of haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. FEBS Lett. 323 267–270. [DOI] [PubMed] [Google Scholar]
- ———. 1993b. Refined X-ray structures of haloalkane dehalogenase at pH 6.2 and pH 8.2 and implications for the reaction mechanism. J. Mol. Biol. 232 856–872. [DOI] [PubMed] [Google Scholar]
- Vriend, G. 1990. WHAT IF: A molecular modeling and drug design program. J. Mol. Graphics 8 52–56. [DOI] [PubMed] [Google Scholar]