Abstract
Oxalate decarboxylase (OXDC) from the wood-rotting fungus Flammulina velutipes, which catalyzes the conversion of oxalate to formic acid and CO2 in a single-step reaction, is a duplicated double-domain germin family enzyme. It has agricultural as well as therapeutic importance. We reported earlier the purification and molecular cloning of OXDC. Knowledge-based modeling of the enzyme reveals a β-barrel core in each of the two domains organized in the hexameric state. A cluster of three histidines suitably juxtaposed to coordinate a divalent metal ion exists in both the domains. Involvement of the two histidine clusters in the catalytic mechanism of the enzyme, possibly through coordination of a metal cofactor, has been hypothesized because all histidine knockout mutants showed total loss of decarboxylase activity. The atomic absorption spectroscopy analysis showed that OXDC contains Mn2+ at up to 2.5 atoms per subunit. Docking of the oxalate in the active site indicates a similar electrostatic environment around the substrate-binding site in the two domains. We suggest that the histidine coordinated manganese is critical for substrate recognition and is directly involved in the catalysis of the enzyme.
Keywords: Oxalate decarboxylase, ECM protein, germin motif, knowledge-based modeling, knockout mutants
Oxalates in animals including humans originate mostly from the diet, especially through ingestion of leafy vegetables and other plant parts. The catabolic pathway of oxalic acid is present in bacteria, fungi, and plants but not in vertebrates. Therefore, management of oxalate in humans is very important because of the absence of any enzyme system that can degrade oxalate. Excess ingestion of oxalate leads to an acute oxalate toxicity, which can result in a variety of disorders including renal failure and urolithiasis (Curhan 1997).
Oxalic acid is widely distributed as calcium and magnesium salts and is catabolized by two major pathways, oxidation (oxalic acid + O2 → 2CO2 + H2O2) and decarboxylation (oxalic acid → formic acid + CO2). The decarboxylation occurs either by activation of oxalic acid to oxalyl CoA that is then degraded by oxalyl CoA decarboxylase (OXAOXA) or directly decarboxylated to formate and CO2. A number of bacterial species, for example, Pseudomonas oxalaticus and Oxalobacter formigenes degrade the oxalate by activation pathway (Chandra and Shethna 1977; Baetz and Allison 1989) which requires ATP, CoA, Mg2+, thiamine pyrophosphate (TPP), and acetate. Oxalate oxidase (OXOX) that converts oxalate to CO2 and H2O2 has been reported from moss (Datta and Meeuse 1955) and other higher plants (Chiriboga 1966; Pundir and Pundir 1993). In contrast, the decarboxylases that catabolize oxalate directly to formate and CO2 have been reported mostly from fungi such as Aspergillus niger (Emiliani and Bekes 1964), Sclerotinia sclerotiorum (Magro et al. 1988), Flammulina velutipes (Mehta and Datta 1991), and Postia placenta (Micales 1997) and at least one from a bacterium, Bacillus subtilis (Tanner and Bornemann 2000). Of four fungal enzymes reported to date, possibly the best characterized is that from the wood-rotting fungus F. velutipes (previously known as Collybia velutipes). All of these decarboxylases are induced by oxalate with the sole exception of B. subtilis enzyme. Bacterial decarboxylase also differs from F. velutipes OXDC in subunit size and pH optimum. The lack of induction of the OXDC ortholog in B. subtilis by exogenous oxalate suggests a role different from that in fungi. It is suggested that the B. subtilis enzyme is involved in decarboxylative phosphorylation similar to that in the gram-negative bacterium O. formigenes, in which the antiporting of oxalate and formate are coupled to oxalate decarboxylation to generate a proton-motive gradient (Maloney 1994). As F. velutipes OXDC is active at low pH (Mehta and Datta 1991) and most of the oxalates reside in plant cell vacuoles, it is conceivable that this enzyme could be targeted to this organelle to generate oxalate-free transgenic plants. OXDC has great agricultural importance as it has been shown to confer improved tolerance in transgenic crops to fungal pathogens such as S. sclerotiorum that use oxalic acid (Kesarwani et al. 2000). OXDC also has great prospects in gene therapy for lowering the oxalate levels in plasma and subsequently in the urine of individuals susceptible to urolithiasis.
The structural basis of the mechanism of OXDC action has not yet been elucidated. OXDC shares the germin motif with two vicilin seed storage proteins. Although the actual sequence homology is not very high, it has been suggested that the structural fold of most of the proteins containing the germin motif is similar (Dunwell and Gane 1997). In this paper, we suggest a model structure of OXDC using phaseolin and canavalin as templates. Experiments designed on the basis of this model led to the identification of a divalent manganese ion bound to a cluster of histidines at the active site of the enzyme. Site-directed mutagenesis involving these histidines suggested that each one of them is critical for enzyme activity. We also propose a geometry of oxalic acid binding to OXDC through the interactions involving manganese ion.
Results
Structure of OXDC
A BLAST search using OXDC sequence with the PDB entries did not yield any good hits. A sequence comparison shows the alignment of OXDC with other germin family proteins (Fig. 1 ▶). In addition to the critical residues within the germin motif, certain residues in other regions are also conserved across these proteins. Although a large number of proteins consisting of germin motif have been characterized (Dunwell and Gane 1997), the three-dimensional structures of only canavalin (Ko et al. 1993), phaseolin (Lawrence et al. 1994), and oxalate oxidase (Woo et al. 2000) have been reported among the different members of this family. Although the amino acid sequence of OXDC shares only 16.6% identity with canavalin and 7.2% identity with phaseolin, the sequence similarity between the germins and the vicilins is statistically significant (Baumlein et al. 1995). The sequence similarity also implies structural similarity and hence forms the basis for the modeling described in this paper. There are reports of building models based on such motif structures in otherwise very low similarity proteins (Chavali et al. 1997; Gane et al. 1998). The sequence alignment of OXDC was done using germin motif as template identified in all three of the proteins. Like OXDC, phaseolin and canavalin are two-domain proteins but contain the germin motif in only the C-terminal domain. The N-terminal domain was aligned such that the OXDC-germin motif aligns with the sequences of phaseolin and canavalin that correspond to the germin motif in the C-terminal domain. Insertions and deletions were adjusted to optimize the homology. The sequence alignment of OXDC with OXAOXA, OXOX, B. subtilis oxalate decarboxylase, and the hypothetical protein from Synechocystis is also shown in Figure 1 ▶. Except for OXOX, these are all double-domain proteins containing a germin motif in each domain. However, OXAOXA, a double-domain protein, lacks the germin motif in either of the two domains. OXDC shows 14.3%, 30.5%, and 46.7% sequence identity with OXAOXA, Synechocystis, and B. subtilis proteins, respectively. OXOX is a single-domain protein which shows 8% identity with the N-terminal and 11.6% with the C-terminal domain of OXDC.
Fig. 1.
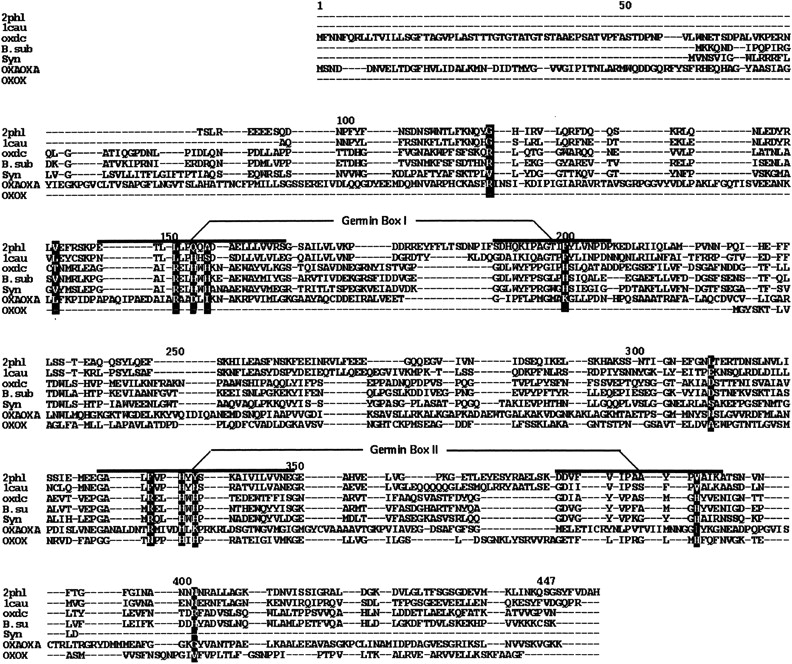
Multiple sequence alignment of OXDC with the other double-domain germin family proteins. Alignment of oxalate-decarboxylase (OXDC) with phaseolin (2phl), canavalin (1cau), oxalate-co A-decarboxylate (OXAOXA), hypothetical proteins from Bacillus subtilis (B. sub) and Synechocystis (Syn). Also shown is the alignment with single-domain oxalate-oxidase (OXOX). The germin box region within each of the domains is marked. The residues of OXDC that interact with oxalate and the corresponding residues from the other proteins have been highlighted.
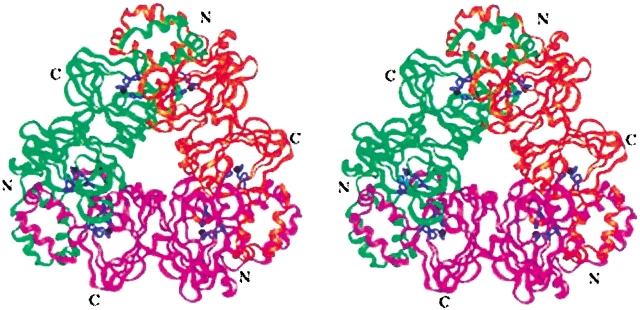
We built a structural model of OXDC on the basis of the sequence alignment using phaseolin and canavalin structures as templates. The first 96 amino acids could not be modeled, as neither of the template protein contains the equivalent region. In any case, this amino terminal segment is not likely to be involved in catalysis, because other cupins (e.g., OXOX) retain the functional activity even though they lack this segment. Assuming that OXDC may exist in the same physiological state as that of phaseolin, a trimer model was constructed using the monomer model and the phaseolin crystal structure. The stereo drawing of the model is shown in Figure 2 ▶. Like phaseolin, OXDC uses α-helical domains to form inter-monomer connections forming an independent four-helix bundle domain involving two α-helices each from the neighboring monomers. Two β-barrel-containing structurally similar domains form the core of the model.
Fig. 2.
Structural model of OXDC. Stereoview of the ribbon drawing showing trimeric protein. Each monomer of the trimer, with two structurally equivalent domains, is shown in a different color. The three histidines in each domain, which appear to juxtapose for metal coordination, are indicated in blue. The possible metal site is shown in black.
The striking feature of OXDC is that the three histidines in each of the two domains (H154, H156, and H199 in the N-terminal domain and H335, H337, and H381 in the C-terminal domain, respectively) that are part of the germin motif form a cluster. The histidines lie on neighboring antiparallel β-strands with two of the histidines in one strand and the third on the adjacent β-strand. The histidines are juxtaposed and thus resemble metal coordinating geometry as observed in metal-binding proteins (Iverson et al. 1990; Yamashita et al. 1990; Pessi et al. 1993; Regan 1995), thereby implying a possible metal binding site within the germin box of OXDC. Interestingly, the histidine cluster in OXDC is similar, structurally, to that of OXOX (Gane et al. 1998). It was recently shown that a single manganese ion is bound per monomer of OXOX by ligands similar to those of manganese superoxide dismutase (Woo et al. 2001). However, neither canavalin nor phaseolin has such histidine triad geometry.
Site-directed mutations in OXDC
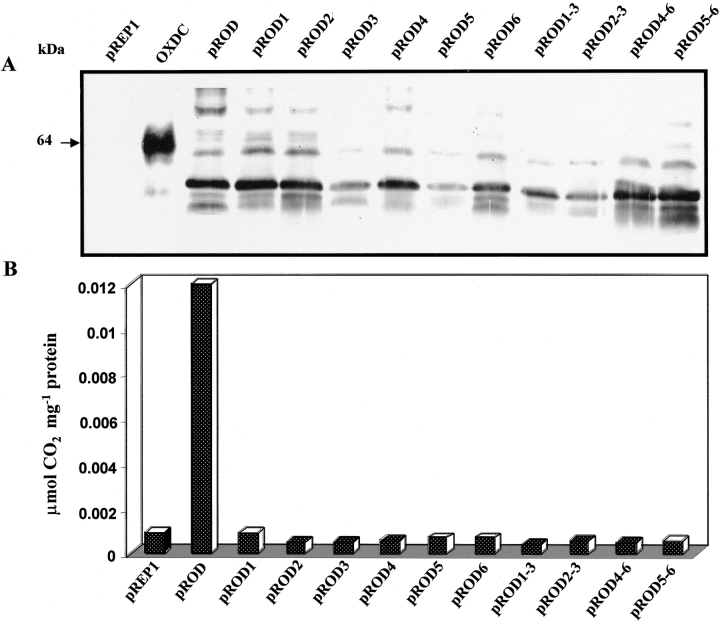
The functional role of the histidine triad in each of the two domains of OXDC monomer was investigated using site-directed mutagenesis. The clone pROD was mutated to incorporate alanine in place of each of the histidines, and six single mutants and four double mutants were generated. As shown in Figure 3A ▶, all of the single as well as the double mutants showed expression of recombinant OXDC protein. The recombinant protein was found to exist in different glycosylated states along with the 64 kD glycosylated form of native OXDC. However, the recombinant protein from pROD was catalytically active. Unlike in the native system, the level of expression of OXDC was very low when expressed in fission yeast. The enzymes thus expressed in fission yeast cells were partially purified at 205-fold with 19.6% recovery. The level of expression in all of the mutants was found to be comparable to that of overexpressed protein in pROD; however, no activity was detected in mutant enzymes (Fig. 3B ▶). These results suggest that all six histidines in the two clusters are critical for OXDC function.
Fig. 3.
Site-directed mutations in OXDC delineate functionally critical histidines. (A) Immunoblot showing the presence of 55-kD OXDC expressed in pROD and all the mutants for histidine. The native OXDC showed a 64-kD band, whereas the vector (pREP1) did not show any signal. The soluble protein fraction (equivalent to 25 μg protein) from the wild type and mutants along with 50 ng of native OXDC was subjected to 10% SDS-PAGE followed by immunoblot analysis using OXDC antibody. The molecular mass of the proteins is indicated. (B) The OXDC activity in the wild type and all mutants was determined as nmole CO2 evolved per mg of protein per min at 37°C. Mean values of triplicate determinations are presented.
Oligomeric organization of OXDC
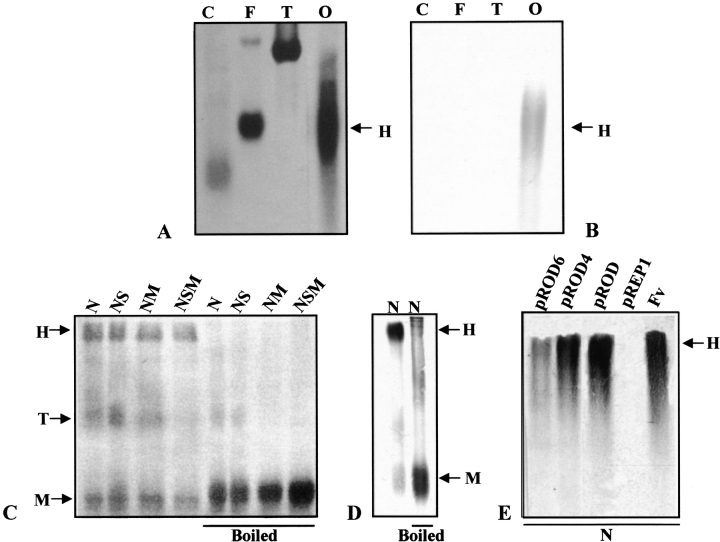
The OXDC from F. velutipes when analyzed by nondenaturing PAGE under nonreducing conditions migrate as a single band with the apparent molecular mass of 420 kD (Fig. 4A,B ▶). Our earlier studies on OXDC using a 7%–15% gradient SDS-PAGE gel showed a single polypeptide of 64 kD, and this molecular size was consistent with all different gel percentages used under reducing conditions (Mehta and Datta 1991), suggesting that OXDC is a hexameric protein. In SDS-PAGE, both under nonreducing and reducing conditions, the protein migrates predominantly as a hexamer with the apparent molecular mass of 410 kD, along with a small amount of trimeric and monomeric forms having the apparent molecular mass of 180 and 64 kD, respectively. When the protein sample is boiled in the presence or absence of a reducing agent (2-Mercaptoethanol) and subjected to SDS-PAGE, it migrates only as a monomeric species (Fig. 4C ▶, lanes 5–8). An immunoblot analysis also revealed the presence of a single monomeric form of OXDC upon boiling in SDS-PAGE under nonreducing conditions (Fig. 4D ▶). Although each subunit of OXDC contains one cysteine residue in the N-terminal domain at 140 positions (Kesarwani et al. 2000; Datta et al. 1996), it is likely that they do not form any intra- or intermolecular disulfide linkages. Thus, the band observed under nonreducing conditions confirms the hexametric form of OXDC with noncovalently linked monomers.
Fig. 4.
Oligomeric organization of OXDC. The purified protein from F. velutipes was subjected to 6% nondenaturating PAGE under nonreducing conditions (A and B) and detected by staining with silver stain (A) and immunoblot analysis using anti-OXDC antibody (B). On 10% SDS-PAGE, the protein was analyzed under different conditions as indicated and detected by silver staining (C) or immunoblot (D). The overexpressed and mutant OXDC were analyzed on 10% SDS-PAGE under native conditions as indicated and detected by immunoblot (E). Monomers (M), trimers (T), and hexamers (H) are indicated by arrows. Lanes: C, catalase; F, ferritin; T, thyroglobulin; O, oxalate decarboxylase; N, native dye; NS, native dye with SDS; NM, native dye with βME; NSM, native dye with SDS and βME; FV, F. velutipes.
The question arises as to whether the oligomerization in recombinant OXDC and/or its mutated forms expressed in fission yeast is identical to that of the native form. To test the event of oligomerization in an alien environment, protein extracts from the clones expressing either recombinant OXDC or its mutated forms were subjected to SDS-PAGE under nonreducing and reducing conditions. The hexameric form of OXDC remained unaltered in pROD as well as mutated clones (Fig. 4E ▶). These results indicate the occurrence of a single noncovalently linked oligomerization of OXDC protomers in all of the clones tested, as in the case of F. velutipes.
Although the homology with phaseolin and canavalin led to the construction of the trimeric structure, the above experimental data indicate that OXDC forms a hexameric assembly. Many enzymes form oligomeric states with a hierarchical organization of subunit assembly (Morera et al. 1994; Engel et al. 1996; Holtham et al. 1999). Thus knowledge-based modeling was used to generate the hexameric structure. Homogentisate dioxygenase (Titus et al. 2000) is a hexamer made up of two trimers akin to the OXDC trimer. The overall shape of each of the monomers of OXDC is similar to that of homogentisate dioxygenase. The OXDC trimer was aligned in sequence with homogentisate dioxygenase, and then the symmetry transformations were applied to the OXDC trimer structure, similar to those of homogentisate dioxygenase, to generate the hexamer (Fig. 5 ▶). The doughnut-shaped molecules stack on top of each other to form the dimmer of trimers.
Fig. 5.
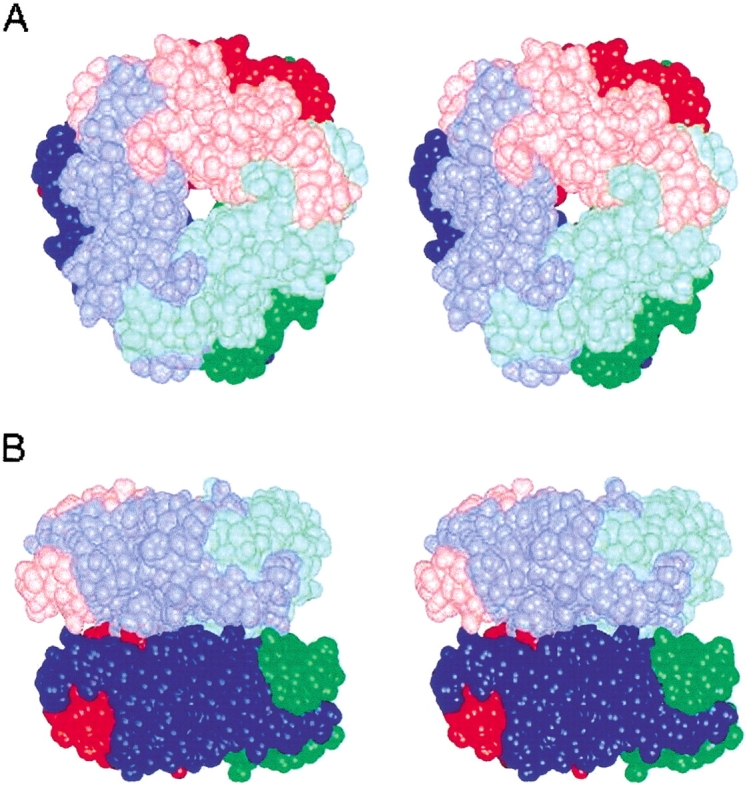
Hexameric assembly of OXDC. Stereoview of space-filling model of the dimer of trimers shown (A) along the threefold axis of the hexamer and (B) along an axis perpendicular to the threefold axis. The three subunits of the trimeric subassembly are shown in red, green, and blue. The top trimer is in light colors, and the bottom trimer is in dark colors.
Presence of bound manganese
The histidine clusters in OXDC are structurally similar to other manganese-binding proteins, leading to the possibility that they define metal-binding sites involving divalent cations such as Cu2+, Mn2+, Zn2+, Ni2+, and Co2+. The presence of divalent cation in purified OXDC from F. velutipes was an6alyzed using atomic absorption spectroscopy. The purification protocol involves two extensive dialysis steps to remove extraneous metal ions. The only metal ion (K+) used during purification of the enzyme was not a candidate cofactor. The results indicated the presence of Mn2+ 1.5 to 2.5 atoms per monomer of OXDC for two different preparations. However, the enzyme samples of both the preparations contained insignificant amounts of Cu, Co, Ni, and Zn (Table 1). Together these data suggest that Mn2+ is tightly bound to OXDC as a cofactor and possibly plays an important role in structural configuration and catalytic activity of the enzyme.
Table 1.
Metal analysis in F. velutipes OXDC
| Number of metal atoms per oxalate decarboxylase monomer | |||||
| Metals | Mn | Cu | Zn | Ni | Co |
| Batch I | 1.80 | 0.12 | 0.08 | 0.09 | 0.10 |
| Batch II | 2.50 | 0.38 | 0.20 | 0.18 | 0.23 |
Presence of divalent cation in OXDC was analyzed by subjecting the purified enzyme from F. velutipes to atomic absorption spectroscopy. Two independent preparations of enzyme were analyzed for identification of divalent cations such as Mn, Cu, Co, Ni, Zn, and Fe. The stoichiometry between the metal ion tested and OXDC was determined as mol/mol and the results expressed as number of atoms per monomer of OXDC.
In an earlier study, we expressed OXDC in Escherichia coli cells grown in Luria-Bertani broth, but the enzyme was functionally inactive (Kesarwani et al. 2000), probably because of incorrect folding of the enzyme. However, in the present study, fission yeast cells could express active OXDC when grown in synthetic medium (EMM) that contains many divalent metal ions including Mn2+, which is otherwise absent from LB broth. The findings in metal analysis showed that the divalent cation Mn2+ is the cofactor of OXDC.
Cooperative binding of oxalic acid to OXDC through histidine cluster
The rate of F. velutipes OXDC activity, when plotted against oxalic acid, showed a sigmoidal response to the increasing substrate concentration. As was observed, in decarboxylation of oxalic acid by OXDC, the interaction coefficient n is >1, suggesting that there are at least two substrate-binding sites per molecule and that the binding of oxalate with OXDC is a cooperative process.
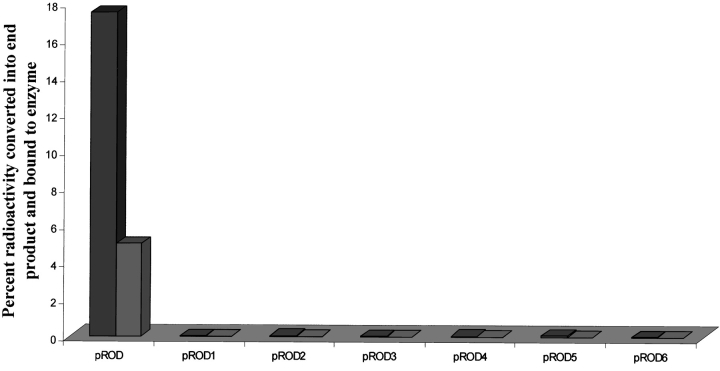
The binding of 14C-labeled oxalic acid to OXDC was determined as described in Materials and Methods. The catalysis of oxalic acid by recombinant OXDC resulted in 17.5% incorporation of radioactivity in the end product with 5% bound to the enzyme-substrate complex in pROD. However, there was no binding of radioactive species either as end product or as intermediate enzyme-substrate complex when mutant enzymes were used (Fig. 6 ▶). The results indicate that none of the mutant enzymes are catalytically active, and the loss of decarboxylation is probably due to alteration in substrate-binding capability. It is very likely that in OXDC, the catalytic and the substrate-binding sites are in the same pocket.
Fig. 6.
Binding of oxalate to recombinant and mutant OXDC expressed in fission yeast. A typical decarboxylation reaction without cold substrate was initiated with 10 nmoles of 14C-oxalic acid and 5 μg equivalent protein extract from recombinant or each independent mutant OXDC. The binding of oxalic acid was determined by percent incorporation of radioactivity in end product (CO2) and remaining reaction mix. The fractions were prepared by trapping CO2, followed by mechanical separation of bound OXDC and free oxalic acid.
Interaction of oxalic acid with OXDC
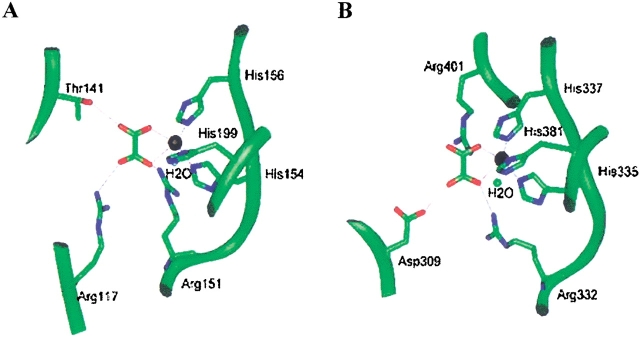
The Mn2+ coordination involving histidines was analyzed in several metal-containing enzyme structures (Tang et al. 1994; Guan et al. 1998; Titus et al. 2000; Woo et al. 2000). The cluster of histidines is present in the central core of the β-barrel in each domain. Structural comparison of the histidine geometry in OXDC structure with that of other existing metal binding proteins shows excellent geometrical fit with three histidine sidechains in OXDC. The manganese was therefore incorporated in the present model at each histidine triad by analogy with the metal-binding sites of other relevant proteins. The model was subjected to further refinement by energy minimization. The proximity of the histidine residues together with the presence of metal ion led us to predict that this must be the potential active site in the molecule. Hence, an attempt was made to dock oxalic acid in the OXDC model to coordinate with the Mn2+ ion using structures of lactoferrin bound to oxalate (PDB-ILCF, Smith et al. 1994) as the template. This was followed by further energy minimization. The oxalic acid shows weaker interaction with OXDC by about 2.0 kcal in the absence of Mn2+. To satisfy the manganese coordination, a water molecule was also included in the model. Modeling of OXDC suggests the possible mode of interaction of the substrate with the enzyme. The arginines, threonine, and aspartic acids come in close proximity to the substrate and therefore might be involved in hydrogen-bonding interactions with the enzyme (Fig. 7A,B ▶).
Fig. 7.
Geometric description of oxalic acid interactions in two domains of OXDC. The interaction of oxalic acid with residues of OXDC and manganese ion in the N-terminal domain (A) and the C-terminal domain (B). The backbone is shown as a ribbon (green) and sidechains as rods. Manganese is shown in black. A water molecule completing metal coordination is also shown. The hydrogen bonds are depicted as thin lines.
Discussion
Several plant storage proteins and other prokaryotic as well as eukaryotic proteins in the germin class apparently exhibit a common structural fold (Argos et al. 1985; Warwicker and O'Connor 1995; Dunwell and Gane 1997; Gane et al. 1998). Sequence analysis of OXDC revealed that it is a double-domain protein that contains a central core sequence called germin motif in each of the two domains. The germin motif is shared by several other functionally diverse enzymes and binding proteins, many of which are associated with synthesis of the extracellular matrix (ECM) (Lane 1994). Certain critical residues within each of these germin motifs are highly conserved, but some residues in other regions are also conserved among all of these proteins (Dunwell and Gane 1997). The germin motif is part of a 20/21 amino acid motif [part of the PRINTS motif, germin-1, G(X)5HXH(X)11G] which is followed usually after 15 residues by a second motif of 16 amino acids [part of the PRINTS motif, germin-2, G(X)5P(X)4H(X)3N]. The size of the intermotif region varies significantly from a minimum of 15 residues in some microbial enzymes to more than 50 residues in the storage proteins and eukaryotic transcription factors (Dunwell et al. 2001). The distance between the two germin boxes in OXDC is 20 amino acids, and the terminal `Asn' in germin box-2 is replaced by an `Ala' residue (Fig. 1 ▶).
We used the sequence information of the germin box family to build a three-dimensional model of OXDC and defined the potential active site geometry based on the above relationship with canavalin and phaseolin in the germin box. The model revealed a hexameric quaternary structure of OXDC molecule organized as a dimer of two trimers, with each monomer containing two structurally similar domains. The buried surface area of the OXDC trimer involved in hexamer interaction is strongly hydrophobic. Approximately 12 aromatic residues appear to be exposed within this area in each trimer. Regarding phaseolin, for which the trimer structure is available, a substantial number of charged residues are exposed in this region, making it a comparatively polar surface. Our experiments using denaturing and nondenaturing PAGE with or without boiling clearly demonstrate the hexameric organization of native OXDC. Moreover, denaturating PAGE under reducing and nonreducing conditions suggests the involvement of noncovalent association among the monomers in each trimer and also between two trimers to form a hexameric structure (Fig. 4A–D ▶). This explains the fact that none of the decarboxylases contain any conserved cysteine that would form a disulfide linkage in hierarchical oligomerization.
The structural model indicated a cluster of three histidine residues, implying a possible metal binding site located within the germin box in the conserved barrel of each of the two domains. The position and orientation of the imidazole rings of the three histidines belonging to the germin box in each of the domains of OXDC resembled ideal topology for the Mn2+ binding site as seen in many other Mn2+-binding proteins (Woo et al. 2000; Guan et al. 1998). In fact, such a topology has been extensively adopted in engineering metal-binding sites in several ab initio and template-based protein design studies (Regan 1995). In the present study, we hypothesized involvement of the two histidine clusters in the catalytic mechanism of the enzyme, possibly through coordination of a metal cofactor, because all of the His → Ala mutants showed total loss of decarboxylase activity. Moreover, noncatalytic vicilin of the cupin superfamily lacks these histidine residues. Indeed, the results of our oxalic acid binding study provide evidence to this effect (Fig. 6 ▶). The cation analysis of the purified enzyme led to the identification of Mn2+ as a dominating species present in the ratio of 2:1 in the purified enzyme. Considering that the substrate of OXDC is an anionic oxalate, the requirement of cation in the anionic substrate-binding pocket might help in tight binding of the other. On the basis of the geometrical arrangements observed in other such enzymes (Banci et al. 1998; Yang et al. 2000), Mn2+ and oxalic acid binding was modeled in both of the domains of OXDC in approximately octahedral coordination of Mn2+. This led to a geometrical arrangement facilitating the coordination requirements of the divalent manganese involving oxalate anion, the nitrogen atoms of the imidazole rings of the three histidines, and a water molecule (Fig. 7 ▶). Other requirements of the manganese environment in the proteins (Guan et al. 1998; Woo et al. 2000) also appear to be reasonably satisfied in both of the domains.
The histidine-coordinated divalent manganese in each of the two domains of OXDC provides scope for the recognition and binding of two oxalate anions in each subunit, completely independent of each other. Yet, the site-specific mutations suggest that knocking out any of the histidine residues in the two clusters leads to complete loss of activity. The possibility that this loss is due to incorrect folding can be ruled out because the mutant proteins showed hexameric assembly in the SDS-PAGE under nondissociating conditions. The allosteric nature of native OXDC suggests this cooperativity between two substrate-binding sites of each monomer. Although the two sites are geometrically independent within a subunit, the active sites of the neighboring subunits are in close proximity in the trimer. In addition to its role in the enzyme catalysis, the metal coordination in proteins also contributes to its architectural stability. Knocking out of any of the histidines should affect the structure of the protein locally, leading to the destabilization of the topology of the other site as well, without hampering the quaternary structure.
The substrate binding sites in the two domains have more or less identical environments in terms of the nature of the residues involved (Fig. 7 ▶). Whereas Arg117 and Thr147 hydrogen-bond with oxalate in the case of the N-terminal domain, the corresponding residues in the C-terminal domain are Arg401 and Asp309. The Arg151 of the N-terminal domain and the corresponding Arg332 of the C-terminal domain form part of the germin motif, and interact with the oxalic acid in both domains. Thus, the electrostatic potential distribution in the substrate-binding pockets of the two domains is comparable but not identical. Such quasiequivalence in the geometry of the active site in two independent domains of a single subunit is interesting.
Comparison of the corresponding residues in other proteins which interact with oxalic acid was also done on the basis of their sequence alignment with OXDC (Fig. 1 ▶). In OXAOXA, the putative substrate-binding non-His residues are conserved, which is consistent with the observation that OXAOXA has catalytic activity similar to that of OXDC. However, OXAOXA does not contain the histidine clusters, implying that it may not have bound metal cation in the substrate-binding pocket. It is attractive to hypothesize that the role of Mn2+ interacting with the substrate in OXDC is assumed by the CoA in the case of OXAOXA. In OXOX, which corresponds to one of the two domains of OXDC, none of the residues except the histidine cluster is conserved. This would explain the difference in the activity of OXOX in comparison to OXDC, whereas the presence of all three histidine residues explains its ion binding property. However, in B. subtilis OXDC, the non-His and His ligands are conserved except for Thr141 and Arg401.
We have demonstrated for the first time the structural model of F. velutipes OXDC consistent with biochemical and molecular evidence for oligomeric organization and histidine-coordinated manganese in substrate recognition. The results suggest that OXDC is a hexamer of six identical subunits of 410 kD molecular mass, and the hexamer is a dimer of trimers. The Mn2+ and oxalic acid binding to the active site of OXDC plays a key role in enzyme catalysis. We also suggest that this motif and histidine may have functional roles in the catalytic mechanism involved in the other two oxalate-degrading enzymes and in the evolution of other proteins with this motif. Our analysis of homologs from diverse prokaryotic and eukaryotic single- or double-domain proteins indicated that, probably, the germin box motif through instruction of divalent cations can give rise to different functions with a corresponding change in the specificity-determining residues.
Materials and methods
Molecular modeling
Knowledge-based modeling of oxalate decarboxylase monomer was carried out using canavalin (PDB code: 1cau) (Ko et al. 1993) and phaseolin (PDB code: 2phl) (Lawrence et al. 1994) as template molecules. The HOMOLOGY module in BIOSYM molecular modeling software (Biosym Technologies) was used on an INDIGO2 workstation (Silicon Graphics) for model building. The structural model was refined by energy minimization in a DISCOVER module. The crystal structures of phaseolin and human homogentisate dioxygenase (PDB code 1EY2) were used to build the trimer and hexamer models, respectively. The sequence alignment of hypothetical proteins from Synechocystis (GenBank accession no. D90907) and B. subtilis oxalate decarboxylase, Yvrk (GenBank acc. no. Z99120 and AF027868) and that of OXAOXA from O. formigenes (GenBank Accession No. M771128) and OXOX from barley (GenBank acc. no. P15290) was accomplished using ClustalW (Thompson et al. 1994) and BLAST (Altschul et al. 1990) searches conducted on the sequences.
Fission yeast strain and growth condition
The fission yeast strain Schizosaccharomyces pombe used in this study was BJ7468 (ura4-D18 leu1-32 ade6-M216). Cells were grown aerobically at 30°C in selective synthetic medium (EMM) as described (Maundrell 1993), and genetic transformation was carried out by the alkaline cation method (Okazaki et al. 1990).
Site-directed mutagenesis
The OXDC coding sequence from a cDNA clone of F. vellutipes (Datta et al. 1996) was subcloned in pREP1 expression vector (Maundrell 1993), and the resulting clone pROD was used as the template for the preparation of site-specific mutants. The mutants pROD1, pROD2, pROD3, pROD4, pROD5, and pROD6 were constructed using the standard PCR-based cloning strategy with mutagenic primers. The double mutants pROD1-3, pROD2-3, pROD4-6, and pROD5-6 were constructed in a similar way using the single mutants as templates. The clones thus obtained were sequenced using the Sequenase™ version 2.0 DNA sequencing kit to rule out the presence of other nonspecific mutations. The DNA templates and the mutagenic oligonucleotides are available upon request.
Purification, expression, and enzyme assay
The OXDC from F. velutipes was purified as described (Mehta and Datta 1991). The overexpressed OXDC from pROD and mutants was extracted in 1 mL of extraction buffer containing 100 mM Tris-Cl (pH 8.0), 1 mM DTT, 20% glycerol, and 3 mM PMSF from the pellet of log-phase (A600 = 3.0) grown cell culture (Rose et al. 1990). The extracts were centrifuged at 12000 g for 5 min. The protein content in supernatant was determined using a Bradford protein assay kit (BioRad). An aliquot of 25 μg enzyme from each sample was subjected to 12.5% SDS-PAGE (Laemmli 1970) and blotted onto a nitrocellulose membrane (Amersham-Pharmacia) by electrotransfer (Towbin et al. 1979). The OXDC enzyme was detected by immunostaining of Western blot using rabbit polyclonal antibody raised against OXDC. The OXDC activity was determined (Mehta and Datta 1991) in 1 mL of reaction mixture using 50 μg equivalent enzyme.
Atomic absorption spectroscopy
The OXDC was purified from F. vellutipes as described (Mehta and Datta 1991), and purified enzyme was analyzed by subjecting the enzyme to Graphite chamber atomic absorption spectrophotometry (Varian Spectra AA 880, GTA 110). The concentration of each metal ion was determined with reference to previously constructed standard curves for all the cations tested.
Substrate binding assays
The substrate-binding assay for F. velutipes OXDC was carried out with purified enzyme, and the assays for pROD and mutant enzymes were done with a partially purified preparation. The decarboxylation reaction was initiated as described (Mehta and Datta 1991) with a few modifications. Each reaction was carried out with only 14C-oxalic acid (10 or 20 nmoles) and 2.5 μg equivalent protein. The radiolabeled CO2 thus evolved was trapped immediately in methylbenzethonium hydroxide and transferred to a separate tube. The reaction was not terminated by TCA but rather was used immediately to separate the protein-bound oxalate from the free oxalate. Thus the remaining reaction was centrifuged through a Centricon-100 filter device at 2000 g for 10 min at 10°C. All three fractions (radiolabeled CO2 as end product, retentate as bound oxalic acid, and flow-through as free one) were transferred to 5 mL of toluene-based scintillation fluid, and the radioactivity was determined. The GenBank accession number for F. velutipes oxalate decarboxylase is AF 200683.
Acknowledgments
This work was supported by grants from the Department of Biotechnology, Government of India.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0206802.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Argos, P., Narayana, S.V.L., and Nielsen, N.C. 1985. Structural similarity between legumin and vicilin storage proteins from legumes. EMBO J. 4 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz, A.L. and Allison, M.J. 1989. Purification and characterization of oxalyl-coenzyme A decarboxylase from Oxalobacter formigenes. J. Bacteriol. 171 2605–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci, L., Bertini, I., Pozzo, L.D., Conte, R.D., and Tien, M. 1998. Monitoring the role of oxalate in manganese peroxidase. Biochemistry 37 9009–9015. [DOI] [PubMed] [Google Scholar]
- Baumlein, H., Braun, H., Kakhovskaya, I.A., and Shutov, A.D. 1995. Seed storage proteins of spermatophytes share a common ancestor with desiccation proteins of fungi. J. Mol. Evol. 41 1070–1075. [DOI] [PubMed] [Google Scholar]
- Chandra, T.S. and Shethna, Y.I. 1977. Oxalate, formate, formamide, and methanol metabolism in Thiobacillus novellas. J. Bacteriol. 131 389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavali, G.B., Nagpal, S., Majumder, S.S., Singh, O., and Salunke, D.M. 1997. Helix-loop-helix motif in GnRH associated peptide is critical for negative regulation of prolactin secretion. J. Mol. Biol. 272 731–740. [DOI] [PubMed] [Google Scholar]
- Chiriboga, J. 1966. Purification and properties of oxalic acid oxidase. Arch. Biochem. Biophys. 116 516–523. [DOI] [PubMed] [Google Scholar]
- Curhan, G.C. 1997. Dietary calcium, dietary protein, and kidney stone formation. Miner. Electrolyte Metab. 23 261–264. [PubMed] [Google Scholar]
- Datta, A., Mehta, A., and Natarajan, K. 1996. Oxalate decarboxylase U.S. patent 5547870.
- Datta, P.K. and Meeuse, B.J.D. 1955. Moss oxalic acid oxidase: A flavoprotein. Biochim. Biophys. Acta 17 602–603. [DOI] [PubMed] [Google Scholar]
- Dunwell, J.M., Culham, A., Carter, C.E., Sosa-Aguirre, C.R., and Goodenough, P.W. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biol. Sci. 26 740–746. [DOI] [PubMed] [Google Scholar]
- Dunwell, J.M. and Gane, P.J. 1997. Microbial relatives of seed storage proteins: Conservation of motifs in a functionally diverse superfamily of enzymes. J. Mol. Evol. 46 147–154. [DOI] [PubMed] [Google Scholar]
- Emiliani, E. and Bekes, P. 1964. Enzymatic oxalate decarborxylation in Aspergillus niger. Arch. Biochem. Biophys. 105 488–493. [DOI] [PubMed] [Google Scholar]
- Engel, C.K., Mathieu, M., Zeelen, J.P., and Wierenga, R.K. 1996. Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 A° resolution: A spiral fold defines the CoA-binding pocket. EMBO J. 15 5135–5145. [PMC free article] [PubMed] [Google Scholar]
- Gane, P.J., Dunwell, J.M., and Warwicker, J. 1998. Modelling based on the structure of vicilins predicts a histidine cluster in the active site of oxalate oxidase. J. Mol. Evol. 46 488–493. [DOI] [PubMed] [Google Scholar]
- Guan, Y., Hickey, M.J., Borgstahl, G.E.O., Hallwell, R.A., Lepock, J.A., O'Connor, D., Hsieh, Y., Nick, H.S., Silverman, D.N., and Tainer, J.A. 1998. Crystal structure of Y34F mutant human mitochondrial manganese superoxide dismutase and the functional role of tyrosine 34. Biochemistry 37 4722–4730. [DOI] [PubMed] [Google Scholar]
- Holtham, C.A.M., Jumel, K., Miller, C M., Harding, S.E., Baumberg, S., and Stockley, P.G. 1999. Probing activation of the prokaryotic arginine transcriptional regulator using chimeric protein. J. Mol. Biol. 289 707–727. [DOI] [PubMed] [Google Scholar]
- Iverson, B.L., Iverson, S.A., Roberts, V.A., Getzoff, E.D., Tainer, J.A., Benkovic, S.J., and Lerner, R.A. 1990. Metalloantibodies. Science 249 659–662. [DOI] [PubMed] [Google Scholar]
- Kesarwani, M., Azam, M., Natarajan, K., Mehta, A., and Datta, A. 2000. Oxalate decarboxylase from Collybia velutipes: Molecular cloning and its overexpression to confer resistance to fungal infection in transgenic tobacco and tomato. J. Biol. Chem. 275 7230–7238. [DOI] [PubMed] [Google Scholar]
- Ko, T.P., Ng, J.D., and McPherson, A. 1993. The three-dimensional structure of canavalin from jack bean (Canavalia ensiformis). Plant Physiol. 101 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lane, B.G. 1994. Oxalate germin and the extracellular matrix of higher plants. FASEB J. 8294–301. [DOI] [PubMed] [Google Scholar]
- Lawrence, M.C., Izard, T., Beuchat, M., Blagrove, R.J., and Colman, P.M. 1994. Structure of phaseolin at 2.2 A° resolution. J. Mol. Biol. 238 748–776. [DOI] [PubMed] [Google Scholar]
- Magro, P., Marciano, P., and Di Lenna, P. 1988. Enzymatic oxalate decarboxylation in isolates of Sclerotinia sclerotiorum. FEMS Lett. 49 49–52. [Google Scholar]
- Maloney, P.C. 1994. Bacterial transporters. Curr. Opin. Cell Biol. 6 571–582. [DOI] [PubMed] [Google Scholar]
- Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123 127–130. [DOI] [PubMed] [Google Scholar]
- Mehta, A. and Datta, A. 1991. Oxalate decarboxylase from Collybia velutipes: Purification, characterization and cDNA cloning. J. Biol. Chem. 266 23548–23553. [PubMed] [Google Scholar]
- Micales, J.A. 1997. Localization and induction of oxalate decarboxylation in the brown-rot wood decay fungus Postia placenta. Int. Biodeterior. Biodegradation 39 125–132. [Google Scholar]
- Morera, S., LeBras, G., Lascu, I., Lacombe, M.L., Veron, M., and Janin, J. 1994. Refined X-ray structure of Dictyostelium discoideum nucleoside diphosphate kinase at 1.8A° resolution. J. Mol. Biol. 243 873–890. [DOI] [PubMed] [Google Scholar]
- Okazaki, K., Okazaki, N., Kume, K., Jinno, S., Tanaka, K., and Okayama, H. 1990. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res. 18 6485–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessi, A., Bianchi, E., Crameri, A., Venturini, S., Tramontano, A., and Sollazzo, M. 1993. A designed metal-binding protein with a novel fold. Nature 362 367–294. [DOI] [PubMed] [Google Scholar]
- Pundir, S. and Pundir, C.S. 1993. Purification and properties of an oxalate oxidase from leaves of grain sorghum hybrid CSH-5. Biochem. Biophys. Acta 1161 1–5. [DOI] [PubMed] [Google Scholar]
- Regan L. 1995. Protein design: Novel metal-binding sites. Trends Biochem. Sci. 20 280–285. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. 1990. Methods in Yeast Genetics: A laboratory course manual. Cold Spring Harbor Laboratory Press, NY, pp. 155–159.
- Smith, C.A., Anderson, B.F., Baker, H.M., and Baker, E.N. 1994. Structure of copper: Substituted and oxalate-substituted human lactoferrin at 2.0 angstrom resolution. Acta Crystallogr.. D50 302. [DOI] [PubMed] [Google Scholar]
- Tang, S-X., Dinner, B.A., Larsen, B.S., Gilchrist, M.L.J., Lorigan, G.A., and Britt, D.R., 1994. Identification of histidine at the catalytic site of the photosynthetic oxygen-evolving complex. Proc. Natl. Acad. Sci. 91 704–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, A. and Bornemann, S. 2000. Bacillus subtilis YvrK is an acid-induced oxalate decarboxylase. J. Bacteriol. 182 5271–5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus, G.P., Mueller, H.A., Burgner, J., de Cordoba, S.R., Penalve, M.A., and Timm, D.E. 2000. Crystal structure of human homogentisate dioxygenase. Nat. Struct. Biol. 7 542–546. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedures and some applications. Proc. Natl. Acad. Sci. 76 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warwicker, J. and O'Connor, J. 1995. A model for vicilin solubility at mild acidic pH, based on homology modeling and electrostatics calculations. Protein Eng. 8 1243–1251 [DOI] [PubMed] [Google Scholar]
- Woo, E-J., Dunwell, J.M., Goodenough, P.W., Marvier, A.C., and Pickersgill, R.W. 2000. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7 1036–1041. [DOI] [PubMed] [Google Scholar]
- Yamashita, M.M., Wesson, L., Eisenman, G., and Eisenberg, D. 1990. Where metal ions bind in proteins. Proc. Natl. Acad. Sci. 87 5648–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Floyd, D.L., Loeber, G., and Tong, L., 2000. Structure of a closed form of human malic enzyme and implications for catalytic mechanism. Nat. Struct. Biol. 7 251–257. [DOI] [PubMed] [Google Scholar]