Abstract
A monomeric form of acetylcholinesterase from the venom of Bungarus fasciatus is converted to a partially unfolded molten globule species by thermal inactivation, and subsequently aggregates rapidly. To separate the kinetics of unfolding from those of aggregation, single molecules of the monomeric enzyme were encapsulated in reverse micelles of Brij 30 in 2,2,4-trimethylpentane, or in large unilamellar vesicles of egg lecithin/cholesterol at various protein/micelle (vesicle) ratios. The first-order rate constant for thermal inactivation at 45°C, of single molecules entrapped within the reverse micelles (0.031 min−1), was higher than in aqueous solution (0.007 min−1) or in the presence of normal micelles (0.020 min−1). This clearly shows that aggregation does not provide the driving force for thermal inactivation of BfAChE. Within the large unilamellar vesicles, at average protein/vesicle ratios of 1:1 and 10:1, the first-order rate constants for thermal inactivation of the encapsulated monomeric acetylcholinesterase, at 53°C, were 0.317 and 0.342 min−1, respectively. A crosslinking technique, utilizing the photosensitive probe, hypericin, showed that thermal denaturation produces a distribution of species ranging from dimers through to large aggregates. Consequently, at a protein/vesicle ratio of 10:1, aggregation can occur upon thermal denaturation. Thus, these experiments also demonstrate that aggregation does not drive the thermal unfolding of Bungarus fasciatus acetylcholinesterase. Our experimental approach also permitted monitoring of recovery of enzymic activity after thermal denaturation in the absence of a competing aggregation process. Whereas no detectable recovery of enzymic activity could be observed in aqueous solution, up to 23% activity could be obtained for enzyme sequestered in the reverse micelles.
Keywords: Acetylcholinesterase, thermal denaturation, protein aggregation, protein unfolding, reverse micelles, large unilamellar vesicles
Protein aggregation is a major medical and biotechnological problem. Diseases such as Alzheimer's disease and other amyloidoses, as well as prion diseases, involve protein aggregation (Sipe 1992; Carrell and Lomas 1997; McParland et al. 2000; Wanker 2000). Aggregation, resulting in the formation of inclusion bodies, is also a common problem in the production of recombinant proteins (Jaenicke 1995; Speed et al. 1996). It is assumed to arise from hydrophobic aggregation of the unfolded or denatured states, resulting in nonregular amorphous oligomeric structures. In contrast, amyloid fibrils arise from native-like conformations, and possess an ordered quaternary structure (Wetzel 1994).
Aggregation of proteins is often considered to occur via nonspecific association of partially folded polypeptide chains through hydrophobic interactions. However, several analyses have suggested that specific intermolecular interactions may be involved (London et al. 1974; Brems et al. 1988; Speed et al. 1996; Yon 1996; Fink 1998; Hammarström et al. 1999), which compete with intramolecular folding to the native state.
In the case of aggregation occurring in the course of protein synthesis, the partially folded molten globule (MG) intermediates arise spontaneously, by collapse of the unfolded polypeptide chains, thus generating the hydrophobic surfaces that lead to aggregation. In the amyloidoses and prion diseases, a substantial conformational transition of the native monomer is a prerequisite for aggregation to occur, and it has been suggested that this may involve partial unfolding to an MG-like intermediate (Safar et al. 1994, 1998).
One issue that arises in analyzing the kinetics and thermodynamics of protein aggregation is the necessity to separate the steps of folding or unfolding of the protein from the aggregation process itself (Mulkerrin and Wetzel 1989; Kendrick et al. 1998). In principle, it is possible that aggregation, which is usually irreversible, could drive unfolding by shifting the equilibrium between partially unfolded and native states. Usually the aggregation pathway, starting from a native protein, is modeled as shown in Scheme 1 ▶, using the Lumry-Eyring treatment. This involves first-order reversible unfolding of the protein, followed by aggregation of the non-native species so generated in higher order processes (Lumry and Eyring 1954; Mulkerrin and Wetzel 1989; Georgiou et al.1994; Plaza del Pino et al. 2000):
Scheme 1.
where N is the native protein, A is an intermediate conformational state preceding aggregation, and Am stands for an aggregated form composed of m protein molecules. This model predicts that aggregation should follow second- or higher order kinetics, and that aggregation should be the driving force in such irreversible protein unfolding, assuming that the aggregation step is rate-limiting.
We earlier showed that a water-soluble acetylcholinesterase dimer, purified from electric organ tissue of Torpedo californica, is converted to a partially unfolded MG state by thermal denaturation (Kreimer et al. 1995). In the presence of dimyristoylphosphatidylcholine liposomes, the rate of inactivation is substantially enhanced (Shin et al. 1997), and the MG dimer generated remains tightly associated with the liposomes (Shin et al. 1996, 1997). More recently, we showed that a monomeric form of AChE, purified from the venom of Bungarus fasciatus (BfAChE) (Cousin et al. 1996b), is similarly converted to a partially unfolded MG by thermal inactivation, and subsequently aggregates rapidly (Shin et al. 1998). Again, the rate of inactivation is substantially enhanced in the presence of dimyristoylphosphatidylcholine liposomes. In this latter case, however, the thermally denatured enzyme does not remain bound to the liposomes, but is released and then aggregates, as is the case for denaturation in the absence of the liposomes. The liposome thus serves as a catalyst of the unfolding process (Shin et al. 1998).
As mentioned above, separation of the folding and aggregation steps is an important issue in analyzing the mechanism(s) of protein denaturation. For this purpose, we adopted a novel approach in which the monomeric BfAChE was encapsulated in either reverse micelles or in large unilamellar phospholipid vesicles (LUV) at various protein/micelle (vesicle) ratios. This permitted us to demonstrate that aggregation does not provide the driving force for the thermal unfolding of BfAChE.
Results
Aggregation in the course of thermal denaturation of BfAChE
We showed earlier (Shin et al. 1998) that under conditions of mild thermal denaturation native BfAChE undergoes a transition to a partially unfolded species with the physicochemical characteristics of a MG (Fink 1995; Ptitsyn 1995), which aggregates rapidly. This is not unexpected, because thermal denaturation of proteins is often accompanied by aggregation, due primarily to the appearance of exposed hydrophobic surfaces in the partially unfolded state (Zale and Klibanov 1986). The rate of thermal inactivation of BfAChE was followed by monitoring the loss of enzymic activity, and the rate of aggregation of the inactivated enzyme was measured by PAGE under nondenaturing conditions. Figure 1A ▶ shows that the rate of aggregation of BfAChE closely resembles the rate of thermal inactivation; Figure 1B ▶ shows the transition from monomer to aggregate on the nondenaturing gel. The sucrose gradient displayed in Figure 2 ▶ confirms that large aggregates are produced by thermal denaturation, in agreement with our earlier observations (Shin et al. 1998).
Fig. 1.
Rates of thermal inactivation and aggregation of BfAChE. Samples of BfAChE (15 μM) in buffer 1 were inactivated at 52°C. Aliquots were withdrawn at appropriate times for assay of enzymic activity or for assessment of monomeric content by PAGE under nondenaturing conditions. (A) Filled circles: residual activity; open triangles: percentage of monomer estimated by scanning of the acrylamide gel. (B) Samples were added to ice-cold sample buffer, frozen, and thawed immediately prior to loading of the acrylamide gel. PAGE under nondenaturing conditions was performed on 5% acrylamide gels as described under Materials and Methods. Each lane was loaded with 10 μg of protein. Staining was with Coomassie blue.
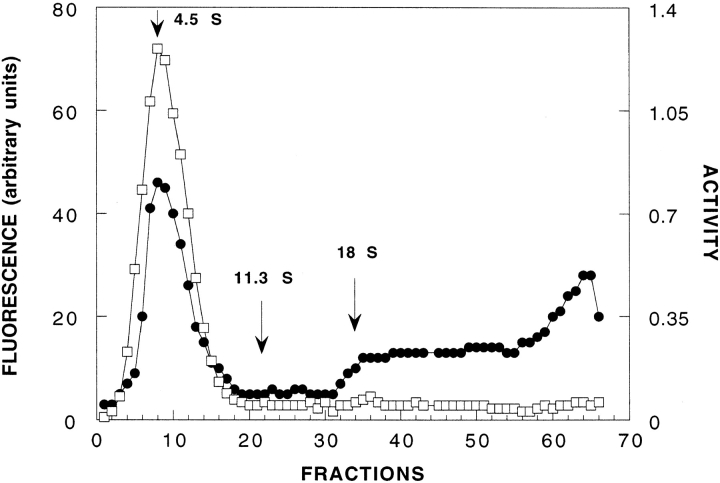
Fig. 2.
Analytical sucrose gradient centrifugation of native and heat-denatured BfAChE. Sucrose gradient centrifugation was performed on 5–20% linear sucrose gradients in buffer 1, essentially as described previously (24). Open squares: native BfAChE; filled circles: BfAChE denatured by heating for 90 sec at 53°C. Native enzyme was assayed by its enzymic activity, and heat-denatured enzyme using intrinsic fluorescence. Catalase (11.3 S), and A12 Electrophorus AChE (18 S), served as internal standards. Centrifugation was performed using an SW40Ti rotor in a Beckman L7–55 ultracentrifuge at 28,000 rpm for 15 h.
The techniques utilized to obtain the data shown in Figures 1 and 2 ▶ ▶ were performed under nondenaturing conditions. To characterize more accurately the molecular species produced by thermal inactivation, we utilized a crosslinking technique that we recently developed (Weiner et al. 1999). This technique takes advantage of the fact that the photosensitive compound, hypericin, preferentially crosslinks MG species of TcAChE, under conditions in which the native dimer is not affected. Figure 3 ▶ displays the results of SDS-PAGE under nonreducing conditions of samples of BfAChE exposed to thermal inactivation at 50°C, followed by irradiation in the presence of hypericin. It can be seen that as inactivation progresses, irradiation produces initially dimers, then heavier oligomers, and eventually large aggregates, which are too heavy to penetrate into the polyacrylamide gel. However, the small oligomeric species produced initially cannot be visualized under nondenaturing conditions, under which they aggregate too rapidly to be "trapped".
Fig. 3.
Crosslinking of native and heat-denatured BfAChE by hypericin as demonstrated by SDS-PAGE. Electrophoresis was performed as described previously, but under nonreducing conditions, using a 3.5–15% acrylamide gradient. Lane 1, native TcAChE dimer (ca. 130 kD). Lane 2, native BfAChE monomer. Lane 3, native BfAChE + hypericin, irradiation for 10 min. Lanes 4–6, heat-denatured BfAChE + hypericin, irradiation for 10 min; heat denaturation was performed for 2, 5, and 10 min, respectively, at 50°C, in buffer 1. Lane M, high molecular weight markers.
Usually, protein aggregation is strongly dependent on both protein concentration and upon temperature, with both collision frequency and hydrophobic interactions driving the association of the partially unfolded species (Zettlmeissl et al. 1979). However, the aggregation of BfAChE, under the conditions of thermal denaturation employed, follows first-order kinetics (k1 = 0.007 min−1), as can be seen from the linearity of plots of ln [monomer] versus time (Fig. 4 ▶). This unexpected result, for a potentially bimolecular reaction, strongly suggests that the rate-limiting step is not that of protein–protein collision but rather the preceding unimolecular step, the conformational transition.
Fig. 4.
Concentration-dependence of the rate of disappearance of monomeric species of BfAChE. The rate of disappearance of monomer, at 53°C, was monitored by PAGE under nondenaturing conditions (see legend to Fig. 1 ▶), and the data were plotted as ln [monomer] versus time. Open circles: 2.5 μM; filled circles: 5 μM; open triangles: 10 μM.
Effect of protein aggregation on the rate of thermal inactivation
Because upon thermal denaturation the rate of aggregation of BfAChE is very similar to the rate of inactivation (Fig. 1 ▶), it is difficult to separate the two processes. One approach that we adopted towards addressing this issue was to encapsulate single molecules of the enzyme in reverse micelles. Entrapment of single macromolecules in vesicles has been used in recent years to approach various biophysical and biochemical problems (see, e.g., Hashimoto et al. 1998; Chiu et al. 1999a, 1999b; Valdez et al. 2001). Recently, Boukobza et al. (2001) developed a system that utilizes surface-tethered lipid vesicles for single biomolecule spectroscopy. This permitted accurate determination of the number of protein molecules in each vesicle from fluorescence time traces. This, in turn, allowed them to demonstrate that the encapsulated molecules display a Poissonian distribution, with, in the particular case analyzed, an average occupancy of 0.65 molecules per vesicle. If we assume a Poissonian distribution for our system, the probability of two protein molecules occupying the same vesicle is two orders of magnitude lower than the probability of occupancy by one molecule.
The reverse micelles were prepared according to Luisi and Steinmann-Hofmann (1987), using Brij 30 in 2,2,4-trimethylpentane, as described under Materials and Methods. To do this, we needed to know both the concentration and size of the reverse micelles so obtained. The surface of one micelle, on the basis of the diameters obtained from the SAXS measurements, is 25,430 Å2 at w0 = 10, and 31,400 Å2 at w0 = 15. The number of molecules of surfactant per vesicle was estimated by assuming that one surfactant molecule occupies 38.5 Å2 (Shimobouji 1989). At the concentration of Brij 30 employed (0.3 M), the concentrations of micelles were calculated to be 4.5 × 10−4 M at w0 = 10, and 3.7 × 10−4 M at w0 = 15. The corresponding concentrations of BfAChE were, respectively, 6.8 × 10−6 and 1.03 × 10−5 M. Thus, in either case, only a small fraction of the reverse micelles, 1.5 and 2.8%, respectively, actually contained a protein molecule.
Utilizing our SAXS data, the volume of the hydrophilic moiety of the reverse micelle is calculated to be 381,500 Å3 at w0 = 10, and 523,000 Å3 at w0 = 15. From analogy with the 3D structure of TcAChE (Sussman et al. 1991), with which it displays high sequence homology and a similar number of amino acids (Cousin et al. 1996a), we can assume that the BfAChE monomer has an ellipsoidal shape, with dimensions ∼45 Å × 60 Å × 65 Å, from which a protein volume of 91,800 Å3 can be calculated. From the number of detergent molecules per micelle, and the volume occupied by each surfactant headgroup, 61.42 Å3 (Suarez et al. 1993), the residual water volume is 219,300 Å3 at w0 = 10, and 322,800 Å3 at w0 = 15. If from these values we subtract the volume of a single protein molecule, viz. 91,800 Å3, we can calculate that at w0 = 10, each protein-containing micelle will contain ∼4200 water molecules, and at w0 = 15, ∼7700 water molecules. Thus, at w0 = 10, the mass of the internal water can be calculated to be only slightly greater than that of the protein, and at w0 = 15, about twofold.
We compared the rates of thermal inactivation of BfAChE inside the reverse micelles, in aqueous solution, and in the presence of normal Brij 30 micelles (Fig. 5 ▶). The first-order rate for inactivation at 45°C within the reverse micelles (0.031 min−1), was higher than both the value observed in aqueous solution (0.007 min−1, not shown), and that observed in the presence of normal micelles at the same concentration of BfAChE, and at a protein/micelle ratio of 1:1 (0.020 min−1). The rate of denaturation in the presence of normal micelles was ∼threefold higher than in their absence, in agreement with our earlier observations (Shin et al. 1998), but both these rates were lower than the rate observed for the enzyme encapsulated in the reverse micelles. This strongly supports the notion that aggregation does not provide the driving force for the thermal inactivation of BfAChE.
Fig. 5.
Kinetics of thermal inactivation of BfAChE inside reverse Brij 30 micelles and in aqueous solution in the presence of normal Brij 30 micelles. Inactivation was performed at 45°C. In both cases, the enzyme concentration was 10 μM. Open circles: BfAChE in reverse Brij 30 micelles, w0 = 15; filled circles: BfAChE in solution in buffer 1 containing 0.1% Brij 30. Monoexponentional fits were obtained, with correlation coefficients of 0.99 and 0.97, respectively, for reverse and normal micelles.
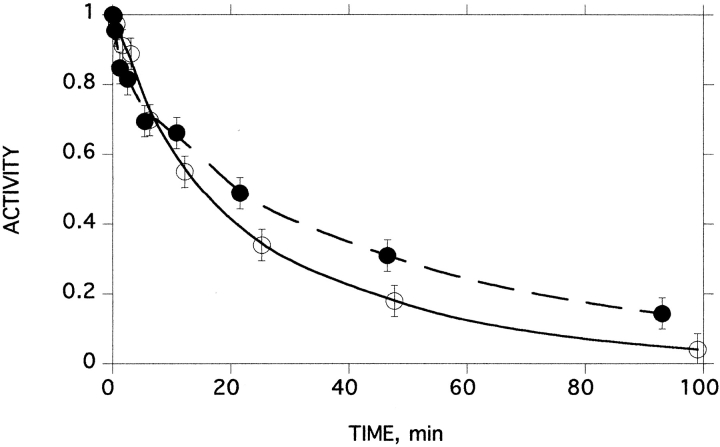
It could be argued that the inner surface of the reverse micelles might enhance the rate of inactivation, thus invalidating our conclusion. Accordingly, we performed experiments in which BfAChE molecules were segregated by entrapment within LUV, at average protein/vesicle ratios of 1:1 and 10:1. The first-order rate constants for inactivation at 53°C were very similar, viz. 0.317 min−1 and 0.342 min−1, respectively (Fig. 6 ▶). As shown above, thermal denaturation produces a distribution of species, ranging from dimers through to large aggregates (Fig. 3 ▶). Consequently, at a protein/vesicle ratio of 10:1, aggregation can occur upon thermal denaturation. Thus, this experiment also supports the notion that aggregation is not a driving force for the thermal unfolding of BfAChE.
Fig. 6.
Kinetics of thermal inactivation of BfAChE inside LUV. Samples of BfAChE entrapped in LUV were incubated at 51°C. Filled circles: protein/vesicle ratio, 1:1; open circles: protein/vesicle ratio, 10:1. Monoexponential fits were obtained with correlation coefficients of 0.99 and 1.00, respectively.
Spectroscopic characteristics of partially unfolded BfAChE
The fact that solutions of reverse micelles of the type employed are optically transparent permits the use of optical spectroscopy for structural characterization of the entrapped protein. We showed earlier that thermally denatured BfAChE displays spectroscopic characteristics typical of a partially unfolded, MG-like state (Shin et al. 1998). To characterize the native and thermally denatured BfAChE within the reverse micelles we utilized CD spectroscopy and intrinsic protein fluorescence. The spectral characteristics of native and of thermally inactivated BfAChE, both in aqueous solution and entrapped within the Brij 30 reverse micelles, are shown in Figures 7 and 8 ▶ ▶. The entrapped BfAChE in reverse micelles appears to retain a structure resembling that of the protein in buffer, as evidenced by the similarity in their spectroscopic properties.
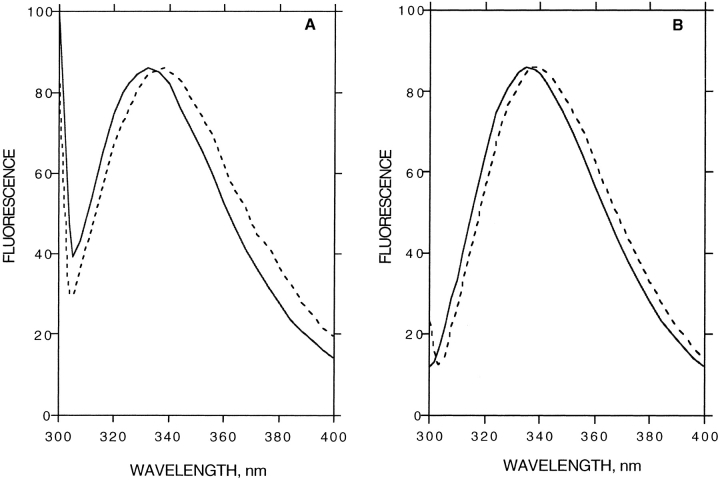
Fig. 7.
Normalized intrinsic fluorescence emission spectra of thermally inactivated BfAChE in reverse micelles and in aqueous solution. BfAChE (2 μM), either entrapped in reverse micelles, w0 = 10 (A), or in aqueous solution (B), was inactivated at 51°C for 15 min. —, native BfAChE; –––, thermally inactivated BfAChE.
Fig. 8.
CD spectra of thermally inactivated BfAChE in reverse micelles and in aqueous solution. BfAChE (8 μM), either entrapped in reverse micelles, w0 = 15 or in aqueous solution, was inactivated at 51°C for 15 min. 1. Native BfAChE; 2. Native BfAChE in reverse micelles; 3. Thermally inactivated BfAChE in aqueous solution; 4. Thermally inactivated BfAChE in reverse micelles.
The intrinsic fluorescence spectrum of BfAChE in buffer has a maximum at 336 nm. The fluorescence emission spectrum of BfAChE in reverse micelles shows a blue shift (about 3 nm) compared with the corresponding spectrum in buffer (Fig. 7 ▶). The blue shift observed in the emission maxima of other proteins and peptides trapped in reverse micelles has been taken to indicate that the entrapped water molecules are somewhat restricted, reducing their capacity for orientational relaxation around the excited-state dipoles of the fluorophores (Bhattacharyya and Basak 1993; Shastry and Eftink 1996). An alternative explanation would ascribe this blue shift to an influence of the hydrophobic moiety of the detergent and of the cosolvent on the excited state of the chromophore (see Luisi and Steinmann-Hofmann 1987).
Thermally denatured BfAChE within the reverse micelles appears to be partially unfolded, because the intrinsic fluorescence emission spectra has a red shift from 333 to 338 nm in the reverse micelles (Fig. 7A ▶) compared to a shift from 336 to 341 nm in buffer (Fig. 7B ▶).
The CD spectra of native and thermally denatured BfAChE in the far UV, within the reverse micelles, are compared to the corresponding spectra in buffer in Figure 8 ▶. The spectra for both the native and denatured BfAChE in the reverse micelles display are very similar to the corresponding spectra in buffer, but a small blue shift, 2–3 nm, can be seen. It should be noted that although the thermally inactivated BfAChE in buffer was aggregated, that in the reverse micelles was not, due to the fact that only single protein molecules were sequestered. Thus, the overall native structure of BfAChE is well preserved in the reverse micelles, although some minor rearrangements cannot be ruled out. Under denaturing conditions, the aggregated form retains a large amount of residual secondary structure, just like the corresponding nonaggregated protein within the reverse micelle. This might, perhaps, be anticipated, because evidence has been presented that proteins display a significant amount of secondary structure within exclusion bodies (Speed et al. 1996).
Refolding of denatured BfAChE
Refolding of a protein from a denatured state to its unique native conformation is limited by competition with aggregation (Mitraki and King 1989). The irreversibility of aggregation underscores the essential role of molecular chaperones in preventing the formation of aggregates in vivo and/or in facilitating the dissociation of multimeric inclusion body intermediates (King et al. 1996). In vitro, molecular chaperones can also protect partially folded intermediates from aggregation and thus promote reactivation (Thirumalai and Lorimer 2001). BfAChE, unfolded in 5 M Gdn HCl, recovered 17% of its original activity when diluted into buffer (Fig. 9 ▶). When dilution was carried similarly, but using the GroEL/GroES chaperone system (Bochkareva et al. 1992) in the presence of MgATP, ∼70% activity was recovered.
Fig. 9.
Time course of refolding of BfAChE unfolded in 5 M Gdn HCl. Unfolding and refolding were as described under Materials and Methods. Refolding was performed in the presence (filled circles) or absence (open circles) of molecular chaperones.
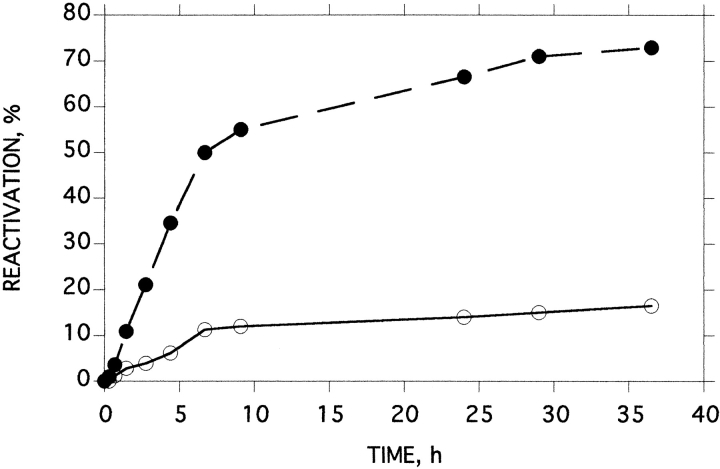
As shown above, another way to protect partially folded intermediates from aggregation is to use either reverse micelles or LUV, in which single protein molecules could be trapped. We tried to reactivate thermally denatured BfAChE under such conditions. When enzyme trapped in reverse micelles was inactivated to ∼4.5% residual activity, by thermal denaturation at 48°C, recovery to 23% was observed upon subsequent cooling, whereas no recovery at all could be detected for enzymes that had been similarly treated in buffer (Fig. 10 ▶). Even though recovery was not quantitative, the experiment clearly showed that such an approach can circumvent the irreversible deactivation due to aggregation.
Fig. 10.
Time course of reactivation of BfAChE after thermal inactivation. Samples of BfAChE in buffer 1, or entrapped inside reverse micelles, w0 = 10, were inactivated at 48°C. When residual activity had reached ∼4.5%, samples were transferred to 4°C, and then slowly reheated to 34°C at a rate of ∼7.5°/h. Open circles: 0.6 μM BfAChE in aqueous buffer; filled circles: entrapped in reverse micelles.
Discussion
Because aggregation is the major cause behind low renaturation yields, elucidating the aggregation pathway may hold the key to successful protein refolding. Intermediates with hydrophobic surfaces exposed to solvent play a crucial role in the partition between native and aggregated conformations. It was suggested earlier that MG intermediates play a major role in the kinetics of folding and aggregation (De Bernardez Clark 1998; Fink 1998; Hammarström et al. 1999). Despite the controversy over the nature of this intermediate (on-pathway versus off-pathway; Creighton 1997) from a kinetic point of view, intermolecular association of MG-like intermediates may indeed be the starting point of the aggregation pathway. Yon (1996), however, has suggested that intermolecular associations responsible for aggregate formation may arise from fluctuating species that precede the MG state.
We showed above that thermally denatured BfAChE undergoes rapid aggregation, and have been able to resolve the processes of denaturation and aggregation by use of either reverse micelles or LUV to segregate single protein monomers. A similar approach was used by Hatton and associates (Hagen et al. 1990) to avoid aggregation in renaturing guanidine-denatured bovine pancreatic ribonuclease A. More recently, they used the same approach, but adopting a solid–liquid extraction technique, to obtain high yields of renatured enzyme (Hashimoto et al. 1998). Reverse micelle systems are claimed to be highly dynamic, even permitting exchange of trapped hydrophilic species via micelle fusion and reseparation (Bru et al. 1995). Nevertheless, the renaturation procedure of Hashimoto et al. (1998), which took place over a period of 30 h, resulted in high yields of renatured protein, even at high protein concentrations, which seems to preclude aggregation via vesicle fusion playing an important role. Thus, it seems reasonable that in our case also, in which renaturation occurred over 5 h, yielding 23% activity, compared to zero activity in the aqueous phase, segregation played an important role in permitting refolding to compete with aggregation. It is, however, possible that vesicle fusion may, nevertheless, contribute to the relatively low yield, although other explanations may be considered.
If it is assumed that a reverse micelle has a fixed volume, then there may also be an effect of volume confinement on the structural state(s) of a protein. As suggested by Minton (1995), macromolecules trapped in a confined volume tend towards their minimum hydrodynamic volume. More explicitly, because an unfolded protein will have less conformational space available to explore inside a reverse micelle, it may be expected that the more compact native state will be stabilized by this confinement effect (Shastry and Eftink 1996). There may, however, also be specific interactions of such a confined protein with the walls of the reverse micelle (Luisi et al. 1988; Nicot and Waks 1996). We, ourselves, have shown that both the surface itself, and surface curvature, play a crucial role in promoting the N→MG transition of both TcAChE and BfAChE during thermal inactivation (Shin et al. 1997, 1998). Because, as shown above, the radius of our reverse micelles is about 50 Å, and the surface to which the trapped protein is exposed possesses negative curvature, it is plausible that the inner surface of the micelle will not increase the rate of the N→MG transition. Moreover, similar data were obtained when small numbers of BfAChE molecules were trapped in LUV, in which confinement was much more limited. In addition, the rates of thermal inactivation of the enzyme trapped inside the reverse micelles was only slightly higher than in solution in the presence of normal micelles (Fig. 5 ▶).
One cannot, however, exclude some form of interaction of the entrapped enzyme with the inner surface of the reverse micelle because, although entrapment of single molecules prevents aggregation, as already pointed out, the yield of renatured enzyme is only ∼23% compared to no detectable reactivation in aqueous solution. As shown above, in the presence of GroEL, GroES, and MgATP, we obtained ∼70% reactivation in free solution, and even in their absence, ∼15%. One of the explanations of this apparent discrepancy is that we are not comparing equivalent experiments. The reactivation experiments within the vesicles are measuring reactivation from a partially unfolded MG state to the native state, with the partially unfolded state being obtained by thermal denaturation. Because denaturation in solution produces immediate aggregation (see Figs. 1–3 ▶ ▶ ▶), reactivation, whether control or chaperone-assisted, involved enzyme samples that had been fully unfolded in Gdn HCl. Thus, the collapsed state obtained upon dilution of the protein in Gdn HCl may not be equivalent to that obtained by unfolding from the native state. It may be noted, also, that for TcAChE we have reported different physicochemical characteristics for two partially unfolded states, one obtained by chemical modification, and the other by exposure to a low concentration of Gdn HCl (Kreimer et al. 1996).
The Lumry-Eyring model shown in Scheme 1 ▶ can be used to describe our data if it is assumed that it is the transition from the native (N) to the aggregation-competent state (A) which is rate-limiting. However, Kendrick et al. (1998), who studied the unfolding of interferon-γ, have suggested a modified scheme in which a transient expansion of the native state (N*), is postulated to precede aggregation, and this led them to modify Scheme 1 ▶ as follows:
Here, the conformational transition of N* to A is the rate-limiting step in the formation of aggregates. They inserted this additional intermediate state to rationalize their observation that the first-order rate constant that they obtained was dependent on sucrose concentration. Despite the fact that the intermediate state (N*) was undetectable for interferon-γ, it is consistent with the irreversibility and first-order kinetics of the aggregation. Earlier experimental data obtained in our laboratory are consistent with such a model, inasmuch as we were able to detect and characterize an intermediate state for Torpedo californica AChE (TcAChE) rather similar to the native state (Kreimer et al. 1994; Shin et al. 1997). Thus, TcAChE in which a buried, nonconserved cysteine, Cys231, has been modified by an organomercurial undergoes a transition to a "quasi-native," N*, state. This quasi-native enzyme, which possesses physicochemical characteristics similar to those of the native enzyme, is devoid of enzymic activity, but displays exposed hydrophobic surfaces, and a somewhat increased hydrodynamic radius. Furthermore, it is metastable, decaying spontaneously, and irreversibly, to a MG state. Despite the fact that such a quasi-native state was not observed experimentally for BfAChE, it is plausible that such a state does indeed exist. In the process of thermal inactivation of BfAChE, the MG state most probably corresponds to A in Scheme 2 ▶. Such an assumption is consistent both with the model of Kendrick et al. (1998) for interferon-γ, and with existence of fluctuating native species that precede the MG state, as suggested by Yon (1996).
Scheme 2.
Our experimental approach, using one or a small number of segregated protein monomers, also permits us to study refolding when the competing aggregation process is excluded. As already mentioned, however, we were not able to obtain more than 23% refolding of thermally denatured BfAChE based on enzymic activity. Non-native intramolecular interactions may stabilize catalytically inactive monomeric species. Indeed, when we used a chaperone system to refold BfAChE unfolded in Gdn HCl, we were able to recover up to 70% enzymic activity. GroEL is thought to assist protein folding in part by providing a passive "box" in which folding can occur without danger of aggregation. However, several recent studies suggest that GroEL not only prevents aggregation, but also plays a more active role by promoting rearrangement, and even unfolding, of kinetic folding intermediates (Coyle et al. 1999; Shtilerman et al. 1999; Wang and Weissman 1999).
Materials and methods
Materials
BfAChE was purified from Bungarus fasciatus venom by affinity chromatography (Cousin et al. 1996b). GroEL and GroES were a gift from Dr. Elena Bochkareva (Department of Biological Chemistry, Weizmann Institute of Science). Hypericin was a gift from Prof. Yehuda Mazur (Department of Organic Chemistry, Weizmann Institute of Science). Tris, 5,5`-dithiobis(2-nitrobenzoic acid), 1-anilino-8-naphthalenesulfonic acid (ANS), egg lecithin, cholesterol, sodium deoxycholate, ATP, dithiothreitol, polyoxyethylene 4-lauryl ether (Brij 30), and high molecular weight markers were from Sigma. Guanidine hydrochloride (Gdn HCl) (ultra pure) was from Schwartz/Mann Biotech. All other reagents were of analytical grade or higher.
Buffers
Unless otherwise stated, the buffer employed was 0.1 M NaCl/10 mM Tris, pH 8.0 (buffer 1).
Assay methods
BfAChE concentrations and activity were determined as described (Dolginova et al. 1992; Shin et al. 1998). Activity of the BfAChE entrapped in the reverse micelles was measured after its extraction from the micelles as follows: One volume of sample was mixed with the same volume of ethyl acetate, and the mixture then centrifuged for 3 min at 12,000 × g. The pellet was dissolved in 200 μL of buffer 1 prior to assay.
Polyacrylamide gel electrophoresis
Polyacrylamide gel electrophoresis (PAGE) under nondenaturing conditions was performed according to (Hames 1990). Sodium-dodecyl-sulfate PAGE was performed as described (Weiner et al. 1999).
Fluorescence measurements
Intrinsic fluorescence of BfAChE and binding of ANS were measured in a Shimadzu RF-540 spectrofluorometer as described earlier (Kreimer et al. 1995).
CD measurements
Circular dichroism (CD) spectra were recorded in a Jasco J-500C spectropolarimeter as described previously (Kreimer et al. 1995). Data are expressed as the mean residue ellipticity, [Θ] (deg cm2 dmol−1).
Light-scattering measurements
The effective hydrodynamic sizes (Dheff) of LUV were measured by quasi-elastic light scattering (QELS) (Berne and Pecora 1976), using an apparatus constructed in-house (Kam et al. 1981). A Spectra Physics 165-4 argon laser was employed with the 514.5-nm line selected. The scattering angle was 90°. Samples were held in borosilicate cells (outer diameter 10 mm), and measurements were performed at 22°C. All samples were filtered before measurement, using 0.22-μm pore diameter Durapore filters (Millipore Corporation). The time autocorrelation function was determined by use of the CONTIN algorithm (Provencher 1979). The viscosity of water at 22°C was taken as 0.96 cP.
Small-angle X-ray scattering
SAXS experiments on Brij 30 reverse micelles in 2,2,4-trimethylpentane were performed using Ni-filtered Cu Kα radiation (0.154 nm) from an Elliott GX6 rotating X-ray generator operating at 1.2 kW. X-irradiation was further monochromated and collimated by a single Franks mirror and a series of slits and height limiters, and detected by a linear position-sensitive detector constructed "in-house". The samples were inserted into 1–1.5-mm quartz or lithium glass capillaries, which were then flame sealed. Each sample was checked after measurement to verify that no fluid had been lost during the time of exposure, ca. 3 h. The temperature was maintained at 45 ± 1°C. The sample-to-detector distance was approximately 0.46 m, and the scattering patterns were desmeared using the procedure of Lake (1967). Micelles were modeled as two-shell spheres consisting of an inner polar core surrounded by a hydrocarbon annulus. Intermicellar interaction was assumed to be "hard-sphere" repulsion, and the corresponding analytical expressions of Sharma and Sharma (1977) were used. Fitting of both the experimental and calculated scattering curves employed Origin (Microcal).
Preparation and characterization of liposomes
LUV with entrapped BfAChE, were prepared using the procedure of Enoch and Strittmatter (1979). Briefly, small unilamellar egg lecithin/cholesterol vesicles (molar ratio 2:1) were prepared by sonication (Shin et al. 1996). The BfAChE to be entrapped was added to the vesicle solution at this point, and the vesicle concentration was adjusted to 20 mM phospholipid. An aliquot of 250 mM sodium deoxycholate was rapidly added and mixed, to yield a final deoxycholate–phospholipid ratio of 1:2. LUV formation was complete within 1 h at 4°C. The bulk of the detergent and free protein were removed by passage over 120 volumes of Sephacryl S-400. The vesicles prepared by this method had an average diameter of 960 ± 120 Å, as determined by QELS, a value similar to that obtained by Enoch and Strittmatter (1979). On this basis, the number of phospholipid molecules per vesicle was calculated to be ∼230,000, using the theoretical curve calculated by Enoch and Strittmatter (1979) for unilamellar egg phosphatidylcholine liposomes.
Preparation and characterization of reverse micelles
The "injection method" (Luisi and Steinmann-Hofmann 1987) was used for solubilization of BfAChE in reverse micelles. Aliquots of either 57 or 85.5 μL of buffer 1, or of 1.2 × 10−4 M BfAChE in buffer 1, were injected into 1 mL of 11.6% Brij 30 in 2,2,4-trimethylpentane. After vortexing for 2 min, the samples were centrifuged for 1 min at 12,000 rpm. Optically clear single-phase solutions were thus obtained, with hydration ratios, w0 = ([H2O]/[Brij 30]), of either 10 or 15.
The size of the reverse micelles was determined by SAXS. Modeling of the SAXS data gave a micelle diameter of 11.6 nm, and a polar core diameter of 9.0 nm for w0 = 10, and of 12.4 and 10.0 nm, respectively, for w0 = 15. The protein had little or no effect on these values.
Unfolding and refolding
Typically, 6 μM BfAChE was unfolded in 5 M Gdn HCl/50 mM Tris, pH 7.2, for 1 h at 22°C. Refolding was initiated by diluting 5 μL of such a solution into 500 μL of either 100 mM KCl/5 mM MgCl2/50 mM Tris, pH 7.2, or into the same buffer containing 10−7M GroEL, 2 × 10−7 M GroES, 5 mM ATP, and 0.1 mM dithiothreitol.
Acknowledgments
This work was supported by a grant from the Israel Science Foundation. I. Silman is the Bernstein-Mason Professor of Neurochemistry, and I. Shin was supported by the Kamea Program for Immigrant Scientists of the Israeli Ministry of Absorption. We thank Elena Bochkareva for advice on the conditions for performing chaperone-assisted refolding experiments, and for supplying GroEL and GroES; Vladimir Budker for advice concerning the protocols for preparation of reverse micelles; Bernard Saliou for preparation of the BfAchE; Yehuda Mazur for providing hypericin; and Gilad Haran, Amnon Horowitz, and Izchak Steinberg for valuable discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
MG, molten globule
N, native
BfAChE, Bungarus fasciatus acetylcholinesterase
LUV, large unilamellar phospholipid vesicles
ANS, 1-anilino-8-naphthalenesulfonic acid
Gdn HCl, guanidine hydrochloride
PAGE, polyacrylamide gel electrophoresis
CD, circular dichroism
QELS, quasi-elastic light scattering
SAXS, small-angle X-ray scattering
TcAchE, Torpedo californica acetylcholinesterase
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0205102.
References
- Berne, B. and Pecora, R. 1976. Dynamic light scattering. Wiley, New York.
- Bhattacharyya, K. and Basak, S. 1993. Fluorescence study of melanocyte stimulating hormones in AOT reverse micelles. Biophys. Chem. 47 21–31. [DOI] [PubMed] [Google Scholar]
- Bochkareva, E.S., Lissin N.M., Flynn, G.C., Rothman, J.E., and Girshovich, A.S. 1992. Positive cooperativity in the functioning of molecular chaperone GroEL. J. Biol. Chem. 267 6796–6800. [PubMed] [Google Scholar]
- Boukobza, E., Sonnenfeld, A., and Haran, G. 2001. Immobilization in surface-tethered lipd vesicles as a new tool for single biomolecule spectroscopy. J. Phys. Chem. B 105 12165–12170. [Google Scholar]
- Brems, D.N., Plaisted, S.M., Havel, H.A., and Tomich, C.S. 1988. Stabilization of an associated folding intermediate of bovine growth hormone by site-directed mutagenesis. Proc. Natl. Acad. Sci. 85 3367–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru, R., Sánchez-Ferrer, A., and Garcia-Carmona, F. 1995. Kinetic models in reverse micelles. Biochem. J. 310 721–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell, R.W. and Lomas, D.A. 1997. Conformational diseases. Lancet 350 134–138. [DOI] [PubMed] [Google Scholar]
- Chiu, D.T., Wilson, C.F., Karlsson, A., Danielsson, A., Lundqvist, A., Strömberg, A., Rytssén, F., Davidson, M., Nordholm, S., Orwar, O., and Zare, R.N. 1999b. Manipulating the biochemical nanoenvironment around single molecules contained within vesicles. Chem. Phys. 247 133–139. [Google Scholar]
- Chiu, D.T., Wilson, C.F., Rytssén, F., Strömberg, A., Farre, C., Karlsson, A., Nordholm, S., Gaggar, A., Modi, B.P., Moscho, A., Garza-Lopéz, R.A., Orwar, O., and Zare, R.N. 1999a. Manipulating the biochemical nanoenvironment around single molecules contained within vesicles. Chemical transformations in individual ultrasmall biomimetic containers. Science 283 1892–1895. [DOI] [PubMed] [Google Scholar]
- Cousin, X., Bon, S., Duval, N., Massoulié, J., and Bon, C. 1996a. Cloning and expression of acetylcholinesterase from Bungarus fasciatus venom. A new type of COOH-terminal domain; Involvement of a positively charged residue in the peripheral site. J. Biol. Chem. 271 15099–15108. [DOI] [PubMed] [Google Scholar]
- Cousin, X., Créminon, Grassi, J., Méflah, K., Cornu, G., Saliou, B., Bon, S., Massoulié, J., and Bon, C. 1996b. Acetylcholinesterase from Bungarus venom: A monomeric species. FEBS Lett. 387 196–200. [DOI] [PubMed] [Google Scholar]
- Coyle, J.E., Texter, F.L., Ashcroft, A.E., Masselos, D., Robinson, C.V., and Radford, S.E. 1999. GroEL accelerates the refolding of hen lysozyme without changing its folding mechanism. Nat. Struct. Biol. 6 683–690. [DOI] [PubMed] [Google Scholar]
- Creighton, T.E. 1997. How important is the molten globule for correct protein folding? Trends Biochem. Sci. 22 6–10. [DOI] [PubMed] [Google Scholar]
- De Bernardez Clark, E. 1998. Refolding of recombinant proteins. Curr. Opin. Biotechnol. 9 157–163. [DOI] [PubMed] [Google Scholar]
- Dolginova, E.A., Roth, E., Silman, I., and Weiner. L. 1992. Chemical modification of Torpedo acetylcholinesterase by disulfides: Appearence of a "molten globule" state. Biochemistry 31 12248–12254. [DOI] [PubMed] [Google Scholar]
- Enoch, H.G. and Strittmatter, P. 1979. Formation and properties of 1000-Å-diameter, single-bilayer phospholipid vesicles. Proc. Natl. Acad. Sci. 76 145–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, A.L. 1995. Compact intermediate states in protein folding. Annu. Rev. Biophys. Biomol. Struct. 24 495–522. [DOI] [PubMed] [Google Scholar]
- Fink, A.L. 1998. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Fold. Des. 3 R9–R23. [DOI] [PubMed] [Google Scholar]
- Georgiou, G., Valax, P., Ostermeier, M., and Horowitz, P.M. 1994. Folding and aggregation of TEM beta-lactamase: Analogies with the formation of inclusion bodies in Escherichia coli. Protein Sci. 3 1953–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen, A.J., Hatton, T.A., and Wang, D.I.C. 1990. Protein refolding in reverse micelles. Biotechnol. Bioeng. 35 955–965. [DOI] [PubMed] [Google Scholar]
- Hames, B.D. 1990. Preparation and electrophoresis of polyacrylamide gels. In Gel electrophoresis of proteins. A practical approach (eds. B.D. Hames and D. Rickwood), pp. 30–50. Oxford University Press, Oxford.
- Hammarström, P., Persson, M., Freskgård, P.O., Mårtensson, L.G., Andersson, D., Jonsson, B.H., and Carlsson, U. 1999. Structural mapping of an aggregation nucleation site in a molten globule intermediate. J. Biol. Chem. 274 32897–32903. [DOI] [PubMed] [Google Scholar]
- Hashimoto, Y., Ono, T., Goto, M., and Hatton, T.A. 1998. Protein refolding by reverse micelles utilizing solid-liquid extraction technique. Biotechnol. Bioeng. 57 620–623. [PubMed] [Google Scholar]
- Jaenicke, R. 1995. Folding and association versus misfolding and aggregation of proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 348 97–105. [DOI] [PubMed] [Google Scholar]
- Kam, Z., Borochov, N., and Eisenberg, H. 1981. Dependence of laser light scattering of DNA on NaCl concentration. Biopolymers 20 2671–2690. [DOI] [PubMed] [Google Scholar]
- Kendrick, B.S., Carpenter, J.F., Cleland, J.L., and Randolph, T.W. 1998. A transient expansion of the native state precedes aggregation of recombinant human interferon-g. Proc. Natl. Acad. Sci. 95 14142–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J., Haase-Pettingell, C., Robinson, A.M., Speed, M., and Mitraki, A. 1996. Thermolabile folding intermediates: Inclusion body precursors and chaperonin substrates. FASEB J. 10 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer, D.I., Dolginova, E.A., Raves, M., Sussman, J.L., Silman, I., and Weiner, L. 1994. A metastable state of Torpedo californica acetylcholinesterase generated by modification with organomercurials. Biochemistry 33 14407–14418. [DOI] [PubMed] [Google Scholar]
- Kreimer, D.I., Shin, I., Shnyrov, V.L., Villar, E., Silman, I., and Weiner, L. 1996. Two partially unfolded states of Torpedo californica acetylcholinesterase. Protein Sci. 5 1852–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreimer, D.I., Shnyrov, V.L., Villar, E., Silman, I., and Weiner, L. 1995. Irreversible thermal denaturation of Torpedo californica acetylcholinesterase. Protein Sci. 4 2349–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake, J.A. 1967. An iterative method of slit-correcting small angle X-ray data. Acta Crystallogr. 23 191–194. [Google Scholar]
- London, J., Skrzynia, C., and Goldberg, M.E. 1974. Renaturation of Escherichia coli tryptophanase after exposure to 8 M urea. Evidence for the existence of nucleation centers. Eur. J. Biochem. 47 409–415. [DOI] [PubMed] [Google Scholar]
- Luisi, P.L. and Steinmann-Hofmann, B. 1987. Activity and conformation of enzymes in reverse micellar solutions. Methods Enzymol. 136 188–229. [DOI] [PubMed] [Google Scholar]
- Luisi, P.L., Giomini, M., Pileni, M.P., and Robinson, B.H. 1988. Reverse micelles as hosts for proteins and small molecules. Biochim. Biophys. Acta 947 209–246. [DOI] [PubMed] [Google Scholar]
- Lumry, R. and Eyring, H. 1954. Conformation changes of protein. J. Phys. Chem. 58 110–120. [Google Scholar]
- McParland, V.J, Kad, N.M., Kalverda, A.P., Brown, A., Kirwin-Jones, P., Hunter, M.G., Sunde, M., and Radford, S.E. 2000. Partially unfolded states of beta(2)-microglobulin and amyloid formation in vitro. Biochemistry 39 8735–8746. [DOI] [PubMed] [Google Scholar]
- Minton, A.P. 1995. Confinement as a determinant of macromolecular structure and reactivity. II. Effects of weakly attractive interactions between confined macrosolutes and confining structures. Biophys. J. 68 1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitraki, A. and King, J. 1989. Protein folding intermediates and inclusion body formation. Bio/Technology 7 690–697. [Google Scholar]
- Mulkerrin, M.G. and Wetzel, R. 1989. pH dependence of the reversible and irreversible thermal denaturation of gamma interferons. Biochemistry 28 6556–6561. [DOI] [PubMed] [Google Scholar]
- Nicot, C. and Waks, M. 1996. Proteins as invited guests of reverse micelles: Conformational effects, significance, applications. Biotechnol. Genet. Eng. Rev. 13 267–314. [DOI] [PubMed] [Google Scholar]
- Plaza del Pino, I.M., Ibarra-Molero, B., and Sanchez-Ruiz, J.M. 2000. Lower kinetic limit to protein thermal stability: A proposal regarding protein stability in vivo and its relation with misfolding diseases. Proteins 40 58–70. [DOI] [PubMed] [Google Scholar]
- Provencher, S.W. 1979. Inverse problems in polymer characterization: Direct analysis of polydispersity with proton correlation spectroscopy. Makromol. Chem. 180 201–209. [Google Scholar]
- Ptitsyn, O.B. 1995. Molten globule and protein folding. Adv. Protein Chem. 47 83–229. [DOI] [PubMed] [Google Scholar]
- Safar, J., Roller, P.P., Gajdusek, D.C., and Gibbs, C.J., Jr. 1994. Scrapie amyloid (prion) protein has the conformational characteristics of an aggregated molten globule folding intermediate. Biochemistry 33 8375–8383. [DOI] [PubMed] [Google Scholar]
- Safar, J., Wille, H., Itri, V., Groth, D., Serban, H., Torchia, M., Cohen, F.E., and Prusiner, S.B. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 4 1157–1165. [DOI] [PubMed] [Google Scholar]
- Shastry, M.C.R. and Eftink, M.R. 1996. Reversible thermal unfolding of ribonuclease T1 in reverse micelles. Biochemistry 35 4094–4101. [DOI] [PubMed] [Google Scholar]
- Shimobouji, T., Matsuoka, H., Ise, N., and Oikawa, H. 1989. Small-angle X-ray scattering studies on nonionic microemulsions. Phys. Rev. A 39 4125–4131. [DOI] [PubMed] [Google Scholar]
- Shin, I., Kreimer, D., Silman, I., and Weiner, L. 1997. Membrane-promoted unfolding of acetylcholinesterase: A possible mechanism for insertion into the lipid bilayer. Proc. Natl. Acad. Sci. 94 2848–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, I., Silman, I., Bon, C., and Weiner, L. 1998. Liposome-catalyzed unfolding of acetylcholinesterase from Bungarus fasciatus. Biochemistry 37 4310–4316. [DOI] [PubMed] [Google Scholar]
- Shin, I., Silman, I., and Weiner, L.M. 1996. Interaction of partially unfolded forms of Torpedo acetylcholinesterase with liposomes. Protein Sci. 5 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtilerman, M., Lorimer, G.H. and Englander, S.W. 1999. Chaperonin function: Folding by forced unfolding. Science 284 822–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe, J.D. 1992. Amyloidosis. Annu. Rev. Biochem. 61 947–975. [DOI] [PubMed] [Google Scholar]
- Speed, M.A., Wang, D.I., and King, J. 1996. Specific aggregation of partially folded polypeptide chains: The molecular basis of inclusion body composition. Nat. Biotechnol. 14 1283–1287. [DOI] [PubMed] [Google Scholar]
- Suarez, M.J., Levy, H., and Lang, J. 1993. Effect of addition of polymer to water-in-oil microemulsions on droplet size and exchange of material between droplets. J. Phys. Chem. 97 9808–9816. [Google Scholar]
- Sussman, J.L., Harel, M., Frolow, F., Oefner, C., Goldman, A., Toker, L., and Silman, I. 1991. Atomic structure of acetylcholinesterase from Torpedo californica: A prototypic acetylcholine-binding protein. Science 253 872–879. [DOI] [PubMed] [Google Scholar]
- Thirumalai, D. and Lorimer, G.H. 2001. Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomol. Struct. 30 245–269. [DOI] [PubMed] [Google Scholar]
- Valdez, D., Le Huérou, J.-Y., Gindre, M., Urbach, W., and Waks, M. 2001. Hydration and water folding in reverse micelles: Compressibility and volume changes. Biophys. J. 80 2751–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J.D. and Weissman, J.S. 1999. Thinking outside the box: New insights into the mechanism of GroEL-mediated protein folding. Nat. Struct. Biol. 6 597–600. [DOI] [PubMed] [Google Scholar]
- Wanker, E.E. 2000. Protein aggregation in Huntington's and Parkinson's disease: Implications for therapy. Mol. Med. Today 6 387–391. [DOI] [PubMed] [Google Scholar]
- Weiner, L., Roth, E., Mazur, Y., and Silman, I. 1999. Targeted cross-linking of a molten globule form of acetylcholinesterase. Biochemistry 38 11401–11405. [DOI] [PubMed] [Google Scholar]
- Wetzel, R. 1994. Mutations and off-pathway aggregation of proteins. Trends Biotechnol. 12 193–198. [DOI] [PubMed] [Google Scholar]
- Yon, J.M. 1996. The specificity of protein aggregation. Nat. Biotechnol. 14 1231. [DOI] [PubMed] [Google Scholar]
- Zale, S.E. and Klibanov, A.M. 1986. Why does ribonuclease irreversibly inactivate at high temperatures? Biochemistry 25 5432–5444. [DOI] [PubMed] [Google Scholar]
- Zettlmeissl, G., Rudolph, R., and Jaenicke, R. 1979. Reconstitution of lactic dehydrogenase. Noncovalent aggregation vs. reactivation. 1. Physical properties and kinetics of aggregation. Biochemistry 18 5567–5571. [DOI] [PubMed] [Google Scholar]