Abstract
The completely sequenced archaeal genomes potentially encode, among their many functionally uncharacterized genes, novel enzymes of biotechnological interest. We have developed a prediction method for detection and classification of enzymes from sequence alone (available at http://www.cbs.dtu.dk/services/ArchaeaFun/). The method does not make use of sequence similarity; rather, it relies on predicted protein features like cotranslational and posttranslational modifications, secondary structure, and simple physical/chemical properties.
Keywords: Function prediction, enzyme classification, Archaea, glycosylation, secondary structure
The conservation among enzymatic pathways is very low in Archaea, and even lower between Bacteria and Archaea. Some of the main characterized pathway operons are not found in some archaea, showing the complete loss of metabolic pathways. This is seen in the case of the histidine pathway that is found in Pyrococcus furiosus but not in Pyrococcus horikoshii or Pyrococcus abyssi (Lecompte et al. 2001). For most pathways, one or two reactions are predicted to be catalyzed by Archaea-specific enzymes (Makarova et al. 1999). As the most studied archaea are all extremophiles, their proteins are of interest to basic science and for commercial exploitation.
In general, the gene repertoire of an archaeal organism is specifically related to other archaea, but is not significantly different from that of bacteria (Bansal and Meyer 2002). The basic components of the transcription, translation, and replication system are well conserved within Archaea, and the same goes for genes involved in repair and recombination.
The archaeal domain of life has a prokaryotic cell organization but is more similar to Eukarya in relation to transcription, translation, and replication. The metabolic proteins in Archaea are often more similar to homologous genes in Bacteria than in Eukarya (Koonin et al. 1997). With the sequencing of the first Crenarchaeota, it was seen that the gene repertoire overlapped more with Euryarchaeota than with Bacteria or Eukarya (Natale et al. 2000). These archaeal features make the archaeal domain of life an interesting area for research in uncharacterized proteins.
The variations in metabolism and the extreme conditions lead to unique archaeal enzymes that need to be characterized. Determination of three-dimensional structure is the traditional approach to functional classification of proteins that cannot be assigned a role based on homology to known proteins. This is a very time-consuming process, and the need for a faster method of classification is obvious. ProtFun is such a method for functional prediction based on sequence-derived features (Jensen et al. 2002); however, it has been developed for eukaryotes.
ProtFun was developed for predicting the function of human proteins and makes use of the fact that the function of a protein is affected by its surroundings and compartment. Functional categories are predicted from correlations between functional features that can be derived from sequence. Some of the features used for human sequences are not biologically meaningful in the case of prokaryotes and thus cannot be expected to correlate with function. However, in a cell without compartments, the function of the protein will still be affected by interacting proteins and cellular components. Interactions between proteins and cellular components can also be derived from the sequence in Archaea. Therefore, we have used the ProtFun approach to make an archaeal enzyme prediction method.
Results and Discussion
Predictability of enzymes and enzyme classes
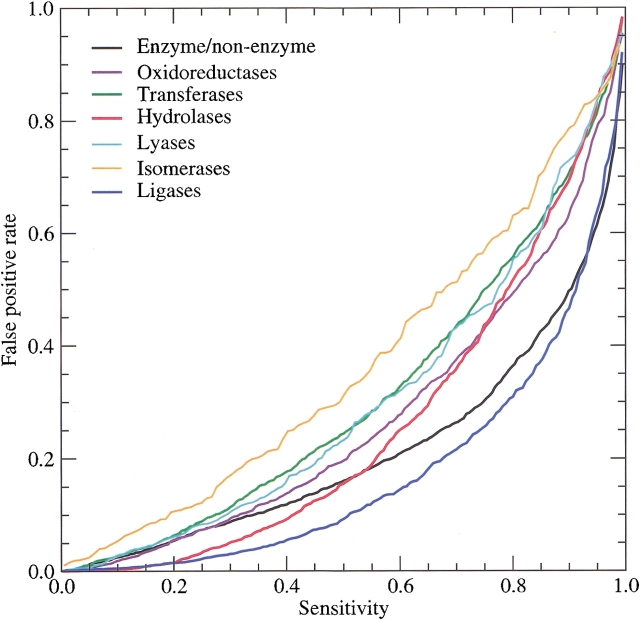
From a practical point of view the most important aspect of a prediction method is its ability to make correct predictions. As prediction methods are never perfect, one is always faced with the dilemma of choosing between making few false-positive predictions and having a high sensitivity, that is, correctly identifying as many positive examples as possible. This tradeoff can be visualized as what is known as the receiver output characteristic (ROC) curve, in which the rate of false positives is plotted as a function of the sensitivity by varying the score threshold used for making positive predictions. Figure 1 ▶ shows the ROC curves for all seven predictors included in our method.
Fig. 1.
Sensitivity and rate of false positives for the different predictors. The plot was constructed based on the results obtained on the cross-validation test set for enzyme/non-enzyme and each of the six major enzyme classes. Owing to the careful partitioning of the cross-validation data set, the performances shown here are what can be expected for real uncharacterized proteins. Random performance corresponds to a line along the diagonal.
The ROC curve for enzyme/non-enzyme prediction breaks at a sensitivity of ∼75% and a false-positive rate of 30%. Assuming that half of the proteins encoded by archaeal genomes are enzymes—which corresponds to the composition of our training set—this corresponds to a specificity just over 70%.
Of proteins that are enzymes, hydrolases and in particular ligases can be predicted with high certainty. For sensitivities below 20%, the rate of false positives is below 1% for both categories. In the case of hydrolases, which are of particular interest in the detergent industry, a sensitivity of 55% can be attained by sacrificing a bit on the specificity. By using the enzyme/non-enzyme and hydrolase predictors in combination, it should thus be possible to predict ∼40% of all novel hydrolases in Archaea.
Isomerases constitute the only class of enzymes in which the performance is too poor for the predictor to be useful for practical purposes. This is somewhat surprising as this enzyme class is, indeed, predictable for human proteins (Jensen et al. 2002). However, the correlations found for human proteins clearly do not carry over to Archaea, as the neural networks trained on human isomerases are not capable of predicting archaeal isomerases either (data not shown). Up to a sensitivity of ∼30%, the remaining enzyme classes are predictable with false-positive rates comparable to those of the enzyme/non-enzyme prediction.
Biologically meaningful feature usage
Although the performance is the most important aspect of the method from an engineering point of view, it is from a scientific point of view at least as interesting to understand how this performance is attained. We now turn to analyzing the biological meaning of the features used for assigning enzymatic function.
Predicted structural properties
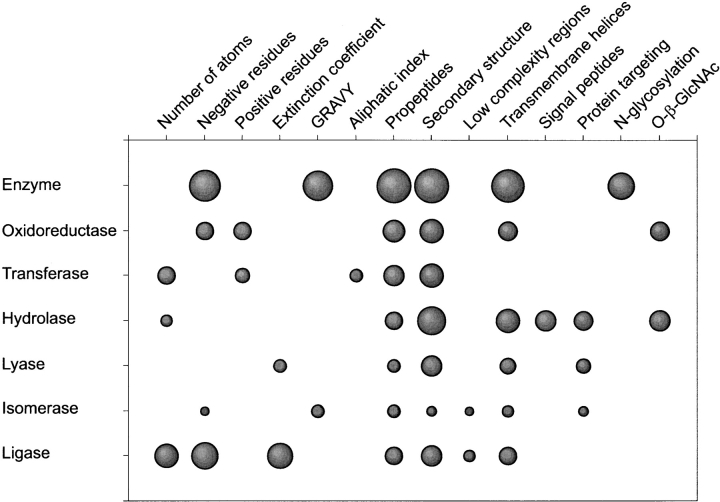
The structure of a protein is an important determinant for the detailed molecular function of proteins, and would consequently also be useful for prediction of enzymes and enzyme classes. As we only have the sequence available, the predicted secondary structure is one of the sequence-derived features that we make use of. Although no clear trends are seen in the secondary structure of different types of enzymes, this feature is one of the most important features for our predictor overall (see Fig. 2 ▶).
Fig. 2.
Feature importance for the different classifiers. The performance of each sequence-derived feature for each category is visualized as a spot with area proportional to the correlation coefficient.
Based on the analysis of secondary structure derived from experimentally determined protein structures, differences in the secondary structure content of enzymes and non-enzymes have previously been shown to exist (Zhang and Zhang 1999). This agrees well with our result that protein secondary structure can be used for predicting enzymes.
Different secondary structure contents have also been observed for particular classes for enzymes, for example, proteases tend to have a lower helix content than other enzymes (Stawiski et al. 2000). Secondary structure is by far the most important feature for prediction of hydrolases, one of the two classes of enzymes that we are best at predicting. By comparing the positional secondary structure content for the six enzyme classes, we found that archaeal hydrolases have an unusually low content of α-helix and high content of β-sheet in their N-terminal region compared with other archaeal enzymes.
One other structural feature that can be predicted from sequence is transmembrane helices. This feature is in particular valuable for prediction enzymes versus non-enzymes, as transmembrane proteins are underrepresented among enzymes. This observation is in sharp contrast to what we have observed for human proteins, where transmembrane helix prediction was found to be useless for enzyme prediction (Jensen et al. 2002). As prokaryotes do not have intracellular membranes, their transmembrane proteins will tend to be distributed over fewer functional categories.
Glycosylation of archaeal proteins
It may come as a surprise that predicted glycosylation sites can be used for the prediction of archaeal enzymes, given that glycosylation is mostly associated with eukaryotes. However, six different types of glycosylation have by now been confirmed to take place in Archaea (Spiro 2002).
We make use of two types of predicted glycosylation sites for this predictor: N-linked β-GlcNAc glycosylation sites are used for the enzyme/non-enzyme prediction, whereas O-β-GlcNAc sites are used for predicting oxidoreductates and hydrolases. The type of N-linked glycosylation is among the six types of glycosylation known to occur in Archaea and is believed to mainly target cell-surface proteins. This may explain why predicted N-glycosylation sites serve as an indicator that a protein is non-enzymatic. The other type of glycosylation that we make use of, O-β-GlcNAc, has not been observed for archaeal proteins so far.
It could be argued that even if the two types of glycosylation sites used in our method exist in Archaea, the predictors cannot be expected to work because they were trained exclusively on eukaryotic data. However, the proteins involved in both types of glycosylation appear to have developed early in evolution, as they are highly conserved among all eukaryotes (Spiro 2002).
In the case of N-linked β-GlcNAc glycosylation, homologs of the highly conserved STT3 subunit of oligosaccharyltransferase have been found, and it has been suggested that the Asn-Xaa-Ser/Thr consensus sequence known in eukaryotes does also hold for archaea (Spiro 2002). Based on this, we find it plausible that the glycosylation predictors, at least NetNglyc, do in fact work for archaea also.
Other important protein properties
In addition to the predicted structural features and glycosylation sites, all predictors make use of one or more of the simple protein properties that can be estimated from the sequence by the ExPASy ProtParam server. This is consistent with observations made in a cross-species validation of the original ProtFun method (data not shown). In particular, it is worth noting that the number of negatively charged residues is usable both for predicting enzymes/non-enzymes and ligases. This is because enzymes in general and ligases in particular contain a high number of negatively charged residues.
The most puzzling feature used in our prediction method is predicted Furin-type propeptide cleavage sites. No such cleavage sites are known in prokaryotic proteins, and the prediction method does not appear to correctly identify archaeal propeptides. In spite of this, the propeptide predictions are correlated to enzyme classes. As Furin-type cleavage sites are mainly characterized by being rich in positively charged residues (arginines and lysines), it is possible that it captures a different simple archaeal signal.
Combining enzyme predictions with phylogenetic patterns
The ProtFun prediction method can be used to obtain functional hints for some of the many archaeal genes for which no functional assignments exist. Additional evidence can be obtained by using information of linked genes based on phylogenetic patterns (Pellegrini et al. 1999). Genes that are linked through a phylogenetic pattern are expected to be within the same cellular role category.
These cellular roles are related to enzyme classes as certain types of enzymes are overrepresented within particular cellular roles. In particular, many proteins involved in energy metabolism are oxidoreductases, whereas central intermediary metabolism is biased toward transferases and hydrolases. Novel proteins from these two cellular role categories were predicted by extracting proteins of unknown function, which in the Predictome database (Mellor et al. 2002) were linked to proteins assigned to cellular roles by EUCLID (Tamames et al. 1998).
Five genes that had only a vague functional description, if any, were found to be linked to proteins involved in central intermediary metabolism and were furthermore predicted by our method to be hydrolases. These proteins had a band-7 domain that is also found in the major integral membrane protein stomatin and in the bacterial plasma membrane proteins HflCK (Tavernarakis et al. 1999). The function of the band-7 domain is unclear. The HflCK proteins have been suggested to be either proteases themselves or modulators of a protease (Cheng et al. 1988; Noble et al. 1993; Kihara et al. 1997). Our prediction agrees with the proposed protease activity.
Many archaeal protein-coding genes have the annotation "conserved hypothetical protein." One such protein from Methanococcus jannaschii, MJ1681p, was linked by a phylogenetic pattern to a number of proteins that were classified as being involved in energy metabolism. Our method predicts MJ1681p to be an oxidoreductase. This prediction is in agreement with Pfam (Bateman et al. 2002), revealing a putative dicluster-type iron–sulfur center. The presence of an iron–sulfur center strongly indicates that this is a ferredoxin, a function that has also been annotated for homologs of MJ1681p in more recently sequenced archaeal genomes.
Materials and methods
Creating labeled data sets
The complete genome sequences of 4 crenarchaea and 10 euryarchaea were downloaded from GenBank (Benson et al. 2002), and the conceptual translations of all 33,143 annotated protein-coding regions were extracted (see Table 1).
Table 1.
The data set size and breakdown on organisms
| Organism | Annotated protein sequences | Assigned as enzyme/non-enzyme | Assigned to enzyme class |
| Aeropyrum pernix | 2694 | 688 | 454 |
| Pyrobaculum aerophilum | 2605 | 863 | 570 |
| Sulfolobus solfataricus | 2977 | 1179 | 782 |
| Sulfolobus tokodaii | 2826 | 1041 | 716 |
| Archaeoglobus fulgidus | 2407 | 1053 | 696 |
| Methanobacterium thermoautotrophicum | 1869 | 878 | 584 |
| Methanococcus jannaschii | 1715 | 780 | 516 |
| Methanococcus mazei | 3371 | 1440 | 908 |
| Methanopyrus kandleri | 1691 | 658 | 455 |
| Methanosarcina acetivorans | 4540 | 1827 | 1123 |
| Pyrococcus abyssi | 1765 | 861 | 541 |
| Pyrococcus horikoshii | 2064 | 781 | 489 |
| Thermoplasma acidophilum | 1031 | 791 | 515 |
| Thermoplasma volcanium | 1499 | 778 | 523 |
| Total | 33,143 | 13,816 | 8872 |
Enzyme classes have been assigned based on sequence similarity to SWISS-PROT entries.
These sequences were searched against the SWISS-PROT database using BLAST (Altschul et al. 1997), recording all matches with an E-value better than 10−3. Using regular expressions, the description lines of all matches were searched for EC numbers, which are annotated for most enzymes in SWISS-PROT. The extracted EC numbers were then used in a majority voting scheme to assign the archaeal query sequences to enzyme/non-enzyme and possibly major enzyme class.
To avoid questionable labeling of the data sets, proteins were only labeled as enzyme if at least two-thirds of their database matches had an EC number in their description line. Equivalently, proteins annotated as non-enzyme were required to have EC numbers for less than one-third of the database matches. If between one-third and two-thirds of the matches had EC numbers or if no matches were found, the classification of the protein was considered unclear and it was removed from the enzyme/non-enzyme data set.
Despite these precautions, the data set will still contain some incorrectly labeled sequences. However, as the function of sufficiently many archaeal proteins has not yet been determined experimentally, one has to rely on function assigned based on sequence similarity.
From proteins annotated as enzyme, a second data set labeled with major enzyme class was assigned based on a similar scheme: If at least two-thirds of the database matches with EC numbers agree on the first digit of the EC number, the query sequence is assigned to the corresponding major enzyme class. As above, sequences were removed if the two-thirds majority rule was not fulfilled.
These procedures resulted in two data sets for each genome: one data set of proteins assigned as either enzyme or non-enzyme, and a smaller set of enzymes annotated with major enzyme class. The sizes of these data sets are shown in Table 1.
Construction of pooled cross-validation sets
Based on an analysis to be presented elsewhere, it was decided to pool the data sets for different organisms to make two large archaeal data sets. As these data sets consist of proteins from different organisms, many orthologous proteins should be expected, making reduction of the similarity between training and test sets particularly important.
The two data sets were each partitioned into five equally sized subsets for cross validation. These sets were constructed with the objective to minimize the total number of significant BLAST hits between sequences in different sets. Each cluster of orthologous proteins will therefore reside in the same subset, allowing for reliable estimation of the performance by cross validation.
Neural network training
Individual cross-validation ensembles of neural networks were trained for predicting the enzyme/non-enzyme classification as well as for predicting each of the six enzyme classes. To avoid problems with overtraining on erroneously labeled examples, networks with very few weights compared with the data set size were used (Brunak et al. 1990). For each of these seven predictors, the optimal combination was found by a boot-strap strategy very similar to that used for the development of the original ProtFun predictor (Jensen et al. 2002).
First a cross-validation training of neural networks was trained, having only a single sequence-derived feature as input. This was done for all features and all categories. The encoding used for each feature can be found at our Web site (http://www.cbs.dtu.dk/services/ProtFun/protfun_add.html). Based on the cross-validated test set performance of these predictors, the worst performing features were rejected for each enzyme category, and neural networks were trained for all pairs of the remaining features. From these the best features were again selected, progressively building up combinations of many features. In the end, the feature combination with the best cross-validation performance was selected for each category.
Prediction on new sequences is done by first running the many prediction methods to obtain the sequence-derived features, which are subsequently used as input for the ensembles of five neural networks for each of the seven categories. The neural network outputs were converted to probability scores as described in the ProtFun publication (Jensen et al. 2002). The probability for each category is estimated as the ensemble average of the probability scores from the individual networks.
Acknowledgments
The authors thank Ramneek Gupta and Peter Duckert for useful discussions on glycosylation and propeptides. This work was supported by grants from the Danish National Research Foundation and the Danish Natural Science Research Council. Marie Skovgaard is funded by EU Cell Factory Project, Screen, QLK3-CT-2000-00649.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.0225102.
References
- Altschul, S., Madden, T., Schaffer, A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal, A. and Meyer, T. 2002. Evolutionary analysis by whole-genome comparisons. J. Bacteriol. 184 2260–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S., Griffiths-Jones, S., Howe, K., Marshall, M., and Sonnhammer, E. 2002. The Pfam protein families database. Nucleic Acids Res. 30 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, D., Karsch-Mizrachi, I., Lipman, D., Ostell, J., Rapp, B., and Wheeler, D. 2002. GenBank. Nucleic Acids Res. 30 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunak, S., Engelbrecht, J., and Knudsen, S. 1990. Cleaning up gene databases. Nature 343 123. [DOI] [PubMed] [Google Scholar]
- Cheng, H., Muhlrad, P., Hoyt, M., and Echols, H. 1988. Cleavage of the cII protein of phage λ by purified HflA protease: Control of the switch between lysis and lysogeny. Proc. Natl. Acad. Sci. 85 7882–7886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, L., Gupta, R., Blom, N., Devos, D., Tamames, J., Kesmir, C., Nielsen, H., Stærfeldt, H., Rapacki, K., Workman, C., Andersen, C., Knudsen, S., Krogh, A., Valencia, A., and Brunak, S. 2002. Prediction of human protein function from post-translational modifications and localization features. J. Mol. Biol. 319 1257–1265. [DOI] [PubMed] [Google Scholar]
- Kihara, A., Akiyama, Y., and Ito, K. 1997. Host regulation of lysogenic decision in bacteriophage λ: Transmembrane modulation of FtsH (HflB), the cII degrading protease, by HflKC (HflA). Proc. Natl. Acad. Sci. 94 5544–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin, E., Mushegian, A., Galperin, M., and Walker, D. 1997. Comparison of archaeal and bacterial genomes: Computer analysis of protein sequences predicts novel functions and suggests a chimeric origin for the archaea. Mol. Microbiol. 25 619–637. [DOI] [PubMed] [Google Scholar]
- Lecompte, O., Ripp, R., Puzos-Barbe, V., Duprat, S., Heilig, R., Dietrich, J., Thierry, J., and Poch, O. 2001. Genome evolution at the genus level: Comparison of three complete genomes of hyperthermophilic archaea. Genome Res. 11 981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova, K., Aravind, L., Galperin, M., Grishin, N., Tatusov, R., Wolf, Y., and Koonin, E. 1999. Comparative genomics of archaea (euryarchaeota): Evolution of conserved protein families, the stable core, and the variable shell. Genome Res. 9 608–628. [PubMed] [Google Scholar]
- Mellor, J., Yanai, I., Clodfelter, K., Mintseis, J., and DeLisi, C. 2002. Predictome: A database of putative functional links between proteins. Nucleic Acids Res. 30 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale, D., Shankavaram, U., Galperin, M., Wolf, Y., Aravind, L., and Koonin, E. 2000. Towards understanding the first genome sequence of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs). Genome Biol. 1 0009.1–0009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, J., Innis, M., Koonin, E., Rudd, K., Banuet, F., and Herskowitz, I. 1993. The Escherichia coli hflA locus encodes a putative GTP-binding protein and two membrane proteins, one of which contains a protease-like domain. Proc. Natl. Acad. Sci. 90 10866–10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, M., Marcotte, E., Thompson, M., Eisenberg, D., and Yeates, T. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 38 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiro, R. 2002. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12 43R–56R. [DOI] [PubMed] [Google Scholar]
- Stawiski, E., Baucom, A., Lohr, S., and Gregoret, L. 2000. Predicting protein function from structure: Unique structural features of proteases. Proc. Natl. Acad. Sci. 97 3954–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamames, J., Ouzounis, C., Casari, G., Sander, C., and Valencia, A. 1998. EUCLID: Automatic classification of proteins in functional classes by their database annotations. Bioinformatics 14 542–543. [DOI] [PubMed] [Google Scholar]
- Tavernarakis, N., Driscoll, M., and Kyrpides, N. 1999. The SPFH domain: Implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24 425–427. [DOI] [PubMed] [Google Scholar]
- Zhang, C.T. and Zhang, R. 1999. Skewed distribution of protein secondary structure contents over the conformational triangle. Prot. Eng. 12 807–809. [DOI] [PubMed] [Google Scholar]